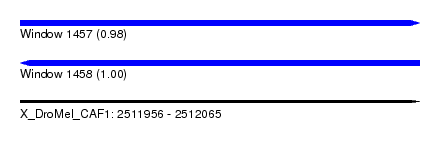
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,511,956 – 2,512,065 |
| Length | 109 |
| Max. P | 0.999851 |

| Location | 2,511,956 – 2,512,065 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 77.48 |
| Mean single sequence MFE | -20.27 |
| Consensus MFE | -13.58 |
| Energy contribution | -13.36 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.983534 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2511956 109 + 22224390 GUUUCUUCUUUUUCGUUAUACAUACAUAUGUAUGUACAUAGUAGAAUAUGAUGUAUGUAUGUAUGAAUAUGUUUGAUAUUUUUAUUUACCACCCCCUGCGUAGGGUGGG ............(((.(((.(((((((((((((((.((((......))))))))))))))))))).)))....)))............((((((........)))))). ( -29.80) >DroSec_CAF1 10424 84 + 1 GUUUCGUUUUAUUU----------------UCUUUCCAUAGUAGA--------UAUGUAUGUAUGAAUAUAUUUG-UAUUUUUAUUUACCACCCCCUGCGUAGGGUGGU ..............----------------...........((((--------(((((........)))))))))-...........(((((((........))))))) ( -15.50) >DroSim_CAF1 11257 84 + 1 GUUUCUUUUUAUUU----------------UCUUUCCAUAGUAGA--------UAUGUAUGUAUGAAUAUGUUUG-UAUUUUUAUUUACCACCCCCUGCGUAGGGUGGU ..............----------------...........((((--------(((((........)))))))))-...........(((((((........))))))) ( -15.50) >consensus GUUUCUUUUUAUUU________________UCUUUCCAUAGUAGA________UAUGUAUGUAUGAAUAUGUUUG_UAUUUUUAUUUACCACCCCCUGCGUAGGGUGGU .......................................((((((.........((((((......))))))........))))))..((((((........)))))). (-13.58 = -13.36 + -0.22)

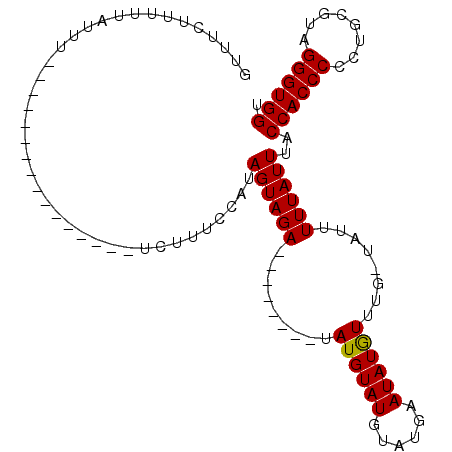
| Location | 2,511,956 – 2,512,065 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 77.48 |
| Mean single sequence MFE | -17.81 |
| Consensus MFE | -12.60 |
| Energy contribution | -12.93 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.49 |
| Structure conservation index | 0.71 |
| SVM decision value | 4.25 |
| SVM RNA-class probability | 0.999851 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2511956 109 - 22224390 CCCACCCUACGCAGGGGGUGGUAAAUAAAAAUAUCAAACAUAUUCAUACAUACAUACAUCAUAUUCUACUAUGUACAUACAUAUGUAUGUAUAACGAAAAAGAAGAAAC .(((((((......)))))))........................(((((((((((...((((......))))........)))))))))))................. ( -23.00) >DroSec_CAF1 10424 84 - 1 ACCACCCUACGCAGGGGGUGGUAAAUAAAAAUA-CAAAUAUAUUCAUACAUACAUA--------UCUACUAUGGAAAGA----------------AAAUAAAACGAAAC ((((((((......))))))))...........-........((((((........--------.....))))))....----------------.............. ( -15.22) >DroSim_CAF1 11257 84 - 1 ACCACCCUACGCAGGGGGUGGUAAAUAAAAAUA-CAAACAUAUUCAUACAUACAUA--------UCUACUAUGGAAAGA----------------AAAUAAAAAGAAAC ((((((((......))))))))...........-........((((((........--------.....))))))....----------------.............. ( -15.22) >consensus ACCACCCUACGCAGGGGGUGGUAAAUAAAAAUA_CAAACAUAUUCAUACAUACAUA________UCUACUAUGGAAAGA________________AAAUAAAAAGAAAC ((((((((......))))))))....................................................................................... (-12.60 = -12.93 + 0.33)

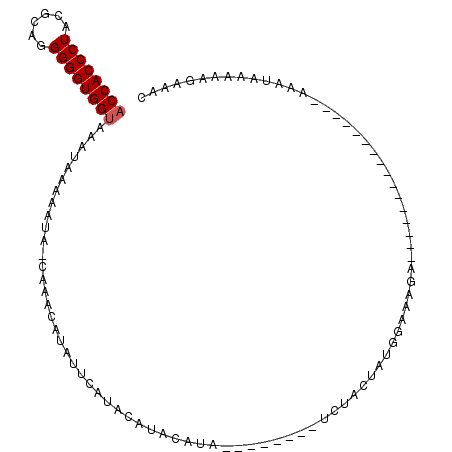
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:56:35 2006