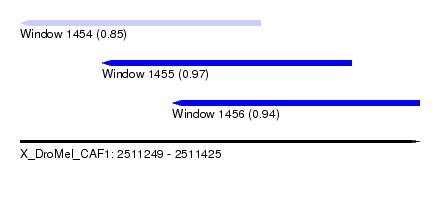
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,511,249 – 2,511,425 |
| Length | 176 |
| Max. P | 0.974785 |

| Location | 2,511,249 – 2,511,355 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.45 |
| Mean single sequence MFE | -17.60 |
| Consensus MFE | -11.04 |
| Energy contribution | -11.28 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.847654 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2511249 106 - 22224390 GAAUGCCCA-ACCCUGGUGGCAACCGCCACCCAAUCACCAAUUACCACCACCACCCACACCU-UUAUAGU------CC----CAAAUGGCAGCAAGAAC-AACCAAAAACUG-GUAAAAA ...((((..-.....((((((....)))))).............................((-....)).------..----.....))))........-.((((.....))-))..... ( -17.80) >DroSec_CAF1 9745 110 - 1 GAAUGCCCA-ACCCUGGUGGCAACCGCCACCCAAUUACCAAUUACCACCA---CCAACACCUUUUCUAGU------CCAAAACAAAUGGCAGCAAGAACCAACCAAAAACUGGGUAAAAA ...((((((-.....((((((....))))))...................---..........((((.((------(((.......)))..)).))))............)))))).... ( -23.10) >DroSim_CAF1 10582 107 - 1 GAAUGCCCA-ACCCUGGUGGCAACCGCCACCCAAUUACCAAUUACCACCA---CCCACACCU-UUCUAGU------CCAAAACAAAUGGCAGCAAGAAC-AACCAAAAACUG-GUAAAAA .........-.....((((((....))))))..........((((((...---.........-((((.((------(((.......)))..)).)))).-..........))-))))... ( -19.40) >DroEre_CAF1 10118 105 - 1 GAAUGUCCAAACACUGGAGGCAACCGCCACCCAAUUACCAAUUACCAC------CCACACCU-UACCCAU------CUAGUACAAAUGGCAGCAAGAAC-AACCAAAAACUG-GUAGAAA ...((.(((...(((((((((....)))....(((.....))).....------........-......)------))))).....)))))........-.((((.....))-))..... ( -13.50) >DroYak_CAF1 11404 110 - 1 GAAUGCCCA-ACCCUGGUGGCAACCACCACCCAAUUACCAAUUCCCAC------CCACACCU-CACCUAUCUAUAUCUAGUUCAAAUGGCAGCAAGAAC-AACCCAAAACUG-GUAGAAA ((((.....-.....(((((......))))).........))))....------........-............((((((((............))))-...(((....))-))))).. ( -14.21) >consensus GAAUGCCCA_ACCCUGGUGGCAACCGCCACCCAAUUACCAAUUACCACCA___CCCACACCU_UUCUAGU______CCAAAACAAAUGGCAGCAAGAAC_AACCAAAAACUG_GUAAAAA ...((((........((((((....)))))).(((.....)))............................................))))............................. (-11.04 = -11.28 + 0.24)

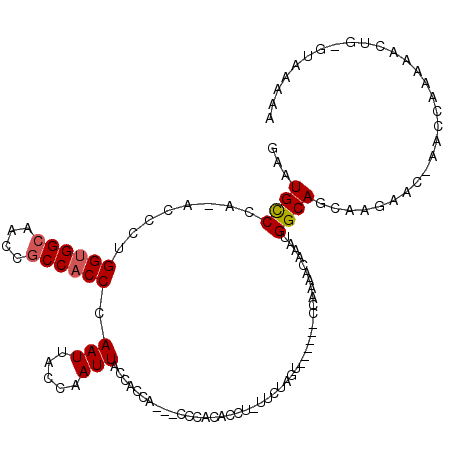
| Location | 2,511,285 – 2,511,395 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.26 |
| Mean single sequence MFE | -27.62 |
| Consensus MFE | -17.52 |
| Energy contribution | -17.88 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.974785 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2511285 110 - 22224390 GAUGGUGGUUAGGUGGUGCCAAAGCCAACACGCGAUUAGUGAAUGCCCA-ACCCUGGUGGCAACCGCCACCCAAUCACCAAUUACCACCACCACCCACACCU-UUAUAGU------CC-- ...((((((..((((((......((......))((((.((((.((....-.....((((((....)))))))).)))).)))))))))))))))).......-.......------..-- ( -35.50) >DroSec_CAF1 9785 102 - 1 --------GGAGGUGGUGCCAAAGCCAACACGCGAUUAGUGAAUGCCCA-ACCCUGGUGGCAACCGCCACCCAAUUACCAAUUACCACCA---CCAACACCUUUUCUAGU------CCAA --------((((((((((.....((...(((.......)))...))...-.....((((((....))))))..............)))))---)).....((.....)))------)).. ( -28.70) >DroSim_CAF1 10620 101 - 1 --------GGAGGUGGUGCCAAAGCCAACACGCGAUUAGUGAAUGCCCA-ACCCUGGUGGCAACCGCCACCCAAUUACCAAUUACCACCA---CCCACACCU-UUCUAGU------CCAA --------((((((((((.....((...(((.......)))...))...-.....((((((....))))))..............)))))---)).....((-....)))------)).. ( -27.80) >DroEre_CAF1 10156 99 - 1 --------GGAAGAGGUGGCAAAGCCAACACGCGAUUAGUGAAUGUCCAAACACUGGAGGCAACCGCCACCCAAUUACCAAUUACCAC------CCACACCU-UACCCAU------CUAG --------((..((((((.....((......))((((.((((.((((((.....))))(((....)))...)).)))).)))).....------...)))))-)..))..------.... ( -23.80) >DroYak_CAF1 11442 104 - 1 --------GGGGAAGGUGCCAAAGCCAACACGCGAUUAGUGAAUGCCCA-ACCCUGGUGGCAACCACCACCCAAUUACCAAUUCCCAC------CCACACCU-CACCUAUCUAUAUCUAG --------(((..(((((.....((...(((.......)))...))...-....((((((...))))))...................------...)))))-..)))............ ( -22.30) >consensus ________GGAGGUGGUGCCAAAGCCAACACGCGAUUAGUGAAUGCCCA_ACCCUGGUGGCAACCGCCACCCAAUUACCAAUUACCACCA___CCCACACCU_UUCUAGU______CCAA ........(((...((((.....((......))((((.((((.((..........((((((....)))))))).)))).))))..............))))...)))............. (-17.52 = -17.88 + 0.36)

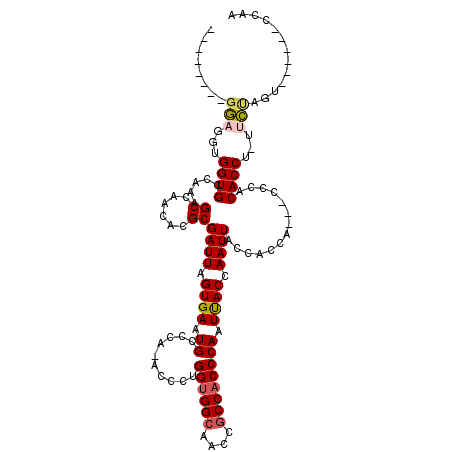
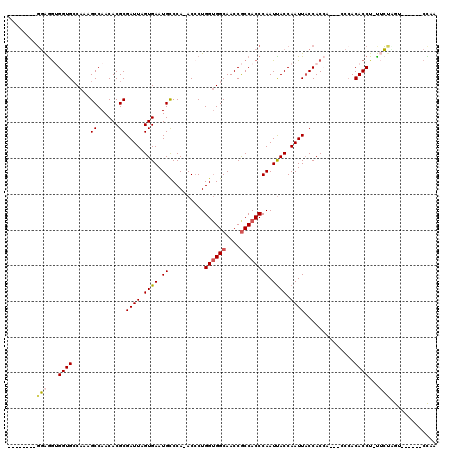
| Location | 2,511,316 – 2,511,425 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 83.41 |
| Mean single sequence MFE | -36.60 |
| Consensus MFE | -27.40 |
| Energy contribution | -27.27 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.941090 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
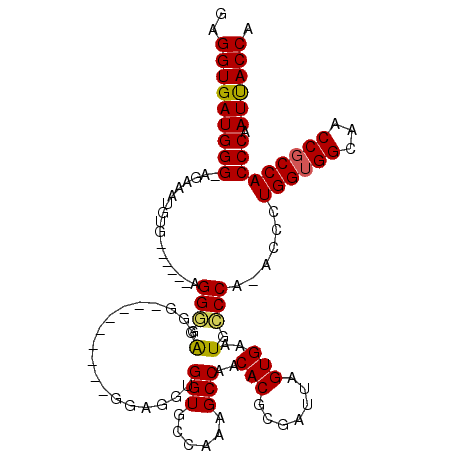
>X_DroMel_CAF1 2511316 109 - 22224390 AAGGUGAUGGG-AGAAAUGUG------AGGGGAGGUAGAUGGUGGUUAGGUGGUGCCAAAGCCAACACGCGAUUAGUGAAUGCCCA-ACCCUGGUGGCAACCGCCACCCAAUCACCA ..(((((((((-.........------((((..((((..((.(((((.((.....))..))))).))(((.....)))..))))..-.))))(((((...))))).))).)))))). ( -40.90) >DroSec_CAF1 9816 99 - 1 GAGGUGAUGGG-AGAAAUGUG------AGGUGAGG----------GGAGGUGGUGCCAAAGCCAACACGCGAUUAGUGAAUGCCCA-ACCCUGGUGGCAACCGCCACCCAAUUACCA ..(((((((((-.........------.(((...(----------((.(.((((......)))).).(((.....)))....))).-))).((((((...))))))))).)))))). ( -34.50) >DroSim_CAF1 10650 99 - 1 GAGGUGAUGGG-GGAAAU-UG------AGGGGAGGG---------GGAGGUGGUGCCAAAGCCAACACGCGAUUAGUGAAUGCCCA-ACCCUGGUGGCAACCGCCACCCAAUUACCA ..((((((.((-(.....-..------((((..(((---------.(.(.((((......)))).).(((.....)))..).))).-.))))(((((...))))).))).)))))). ( -39.40) >DroEre_CAF1 10183 108 - 1 GAGGUGAUGGGGAGAAAUACGAAUACGAGGAGGAGG---------GGAAGAGGUGGCAAAGCCAACACGCGAUUAGUGAAUGUCCAAACACUGGAGGCAACCGCCACCCAAUUACCA ..((((((.(((.......((....)).........---------......(((((....(((..(((.......)))....((((.....)))))))..))))).))).)))))). ( -31.60) >consensus GAGGUGAUGGG_AGAAAUGUG______AGGGGAGGG_________GGAGGUGGUGCCAAAGCCAACACGCGAUUAGUGAAUGCCCA_ACCCUGGUGGCAACCGCCACCCAAUUACCA ..(((((((((.................(((.(..................(((......)))..(((.......)))..).)))......((((((...))))))))).)))))). (-27.40 = -27.27 + -0.12)

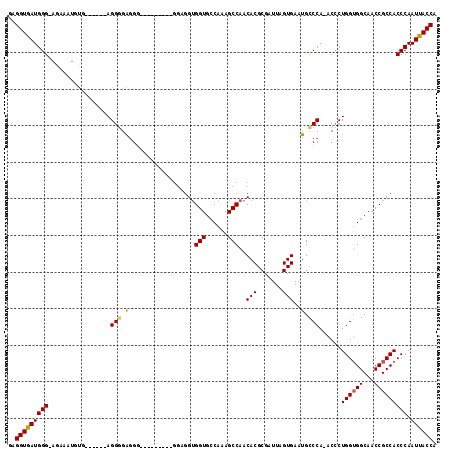
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:56:33 2006