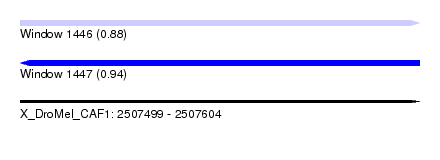
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,507,499 – 2,507,604 |
| Length | 105 |
| Max. P | 0.937237 |

| Location | 2,507,499 – 2,507,604 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.10 |
| Mean single sequence MFE | -23.22 |
| Consensus MFE | -12.00 |
| Energy contribution | -12.40 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.876093 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2507499 105 + 22224390 UCGCCAAAACUUUCCAGUCUGCUGCACAACAAUGUGAAACGGAACACGAAACGUUCCUCCUCUUCUUGAUUCACAUAUGUGUGCAUAU---------------AUAGACCGACAGACAGA .........((.((..(((((.((((((...(((((((..(((((.......)))))...((.....)))))))))...))))))...---------------.))))).)).))..... ( -26.50) >DroSec_CAF1 6073 97 + 1 UCGCCAAAACUUUCCAGUCUGCUGCACAACAAUGUGAAACGGAACACGAAACGUUCC---UCUUCUUGUUUCACAUAUCUGUACAU-----------------AUAGACCGACAGAC--- ................((((((.........((((((((((((((.......)))))---.......))))))))).(((((....-----------------)))))..).)))))--- ( -21.81) >DroSim_CAF1 6979 97 + 1 UCGCCAAAACUUUCCAGUCUGCUGCACAACAAUGUGAAACGGAACACGAAACGUUCC---UCUUCUUGAUUCACAUAUCUGUACAU-----------------AUAGACCGACAGAC--- ................((((((.........(((((((..(((((.......)))))---((.....))))))))).(((((....-----------------)))))..).)))))--- ( -19.80) >DroEre_CAF1 6498 112 + 1 UCGCCAAAACUUUCCAGUCUGCUGCACAACAAUGUGAAACGGAACACGAAACGUUCC---UCUUCUUGAUUCACAUAUAUGUCCAUAUAUC---UAUAUACAUAUUAACAG--UGACAUA ................(((.((((.......(((((((..(((((.......)))))---((.....)))))))))((((((..((((...---.))))))))))...)))--))))... ( -21.50) >DroYak_CAF1 7555 117 + 1 UCGCCAAAACUUUCCAGUCUGCUGCACAACAAUGUGAAACGGAACACGAAACGUUCC---UCUUCUUGAUUCACAUAUGUAUAUAUCUCUCUUGUAUAUACAUAUUGACUGACUUGAAUA ..............(((((.............((((((..(((((.......)))))---((.....))))))))(((((((((((.......)))))))))))..)))))......... ( -26.50) >consensus UCGCCAAAACUUUCCAGUCUGCUGCACAACAAUGUGAAACGGAACACGAAACGUUCC___UCUUCUUGAUUCACAUAUCUGUACAU_U_______________AUAGACCGACAGACA_A ..............(((....))).(((...(((((((..(((((.......)))))...((.....)))))))))...)))...................................... (-12.00 = -12.40 + 0.40)

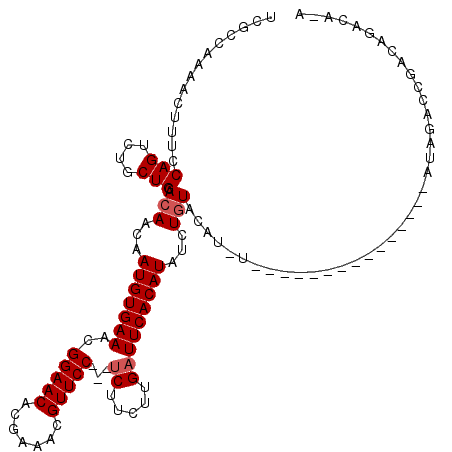
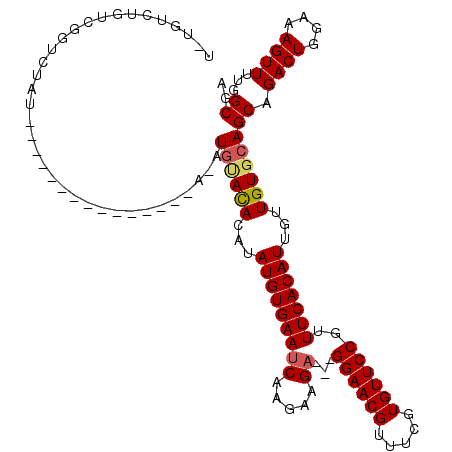
| Location | 2,507,499 – 2,507,604 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.10 |
| Mean single sequence MFE | -30.62 |
| Consensus MFE | -22.92 |
| Energy contribution | -23.24 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937237 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2507499 105 - 22224390 UCUGUCUGUCGGUCUAU---------------AUAUGCACACAUAUGUGAAUCAAGAAGAGGAGGAACGUUUCGUGUUCCGUUUCACAUUGUUGUGCAGCAGACUGGAAAGUUUUGGCGA ..((((..(((((((..---------------...((((((((.(((((((((.....))...((((((.....))))))..))))))))).))))))..)))))))........)))). ( -30.90) >DroSec_CAF1 6073 97 - 1 ---GUCUGUCGGUCUAU-----------------AUGUACAGAUAUGUGAAACAAGAAGA---GGAACGUUUCGUGUUCCGUUUCACAUUGUUGUGCAGCAGACUGGAAAGUUUUGGCGA ---(((..(((((((..-----------------.(((((((..(((((((((.......---((((((.....)))))))))))))))..)))))))..)))))))........))).. ( -32.71) >DroSim_CAF1 6979 97 - 1 ---GUCUGUCGGUCUAU-----------------AUGUACAGAUAUGUGAAUCAAGAAGA---GGAACGUUUCGUGUUCCGUUUCACAUUGUUGUGCAGCAGACUGGAAAGUUUUGGCGA ---(((..(((((((..-----------------.(((((((..(((((((((.....))---((((((.....))))))..)))))))..)))))))..)))))))........))).. ( -30.70) >DroEre_CAF1 6498 112 - 1 UAUGUCA--CUGUUAAUAUGUAUAUA---GAUAUAUGGACAUAUAUGUGAAUCAAGAAGA---GGAACGUUUCGUGUUCCGUUUCACAUUGUUGUGCAGCAGACUGGAAAGUUUUGGCGA ..(((((--(((((.((((((.....---.))))))(.(((...(((((((((.....))---((((((.....))))))..)))))))...))).))))))(((....)))..))))). ( -29.00) >DroYak_CAF1 7555 117 - 1 UAUUCAAGUCAGUCAAUAUGUAUAUACAAGAGAGAUAUAUACAUAUGUGAAUCAAGAAGA---GGAACGUUUCGUGUUCCGUUUCACAUUGUUGUGCAGCAGACUGGAAAGUUUUGGCGA .......((((.(((((((((((((((......).))))))))))).)))..........---((((((.....))))))(((.((((....)))).)))(((((....))))))))).. ( -29.80) >consensus U_UGUCUGUCGGUCUAU_______________A_AUGUACACAUAUGUGAAUCAAGAAGA___GGAACGUUUCGUGUUCCGUUUCACAUUGUUGUGCAGCAGACUGGAAAGUUUUGGCGA ...................................((((((...(((((((((.....))...((((((.....))))))..)))))))...))))))((.((((....))))...)).. (-22.92 = -23.24 + 0.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:56:25 2006