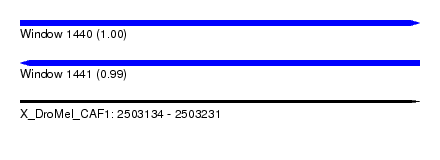
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,503,134 – 2,503,231 |
| Length | 97 |
| Max. P | 0.999451 |

| Location | 2,503,134 – 2,503,231 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.37 |
| Mean single sequence MFE | -28.88 |
| Consensus MFE | -25.46 |
| Energy contribution | -25.38 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.65 |
| Structure conservation index | 0.88 |
| SVM decision value | 3.61 |
| SVM RNA-class probability | 0.999451 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2503134 97 + 22224390 GAAAAUCACGAAAAGA--------GCUAUCAACAU---GGCCUAAAGUGACCAAUGAAUUGGAGUUUAUUUCACGGCGUUGUGAAAUAUGCUCCAACUUUAAC------CC--A----AA .....((((....((.--------(((((....))---)))))...))))....(((((((((((.(((((((((....))))))))).))))))).))))..------..--.----.. ( -27.80) >DroSec_CAF1 1820 97 + 1 GAAAAUCACGAAAAGA--------GCUAUCAACAU---GGCCCAAAGUGGCCAAUGAAUUGGAGUUUAUUUCACGUCGUUGUGAAAUGUGCUCCAACUUUAAC------CC--A----AA ...........((((.--------........(((---((((......)))).)))..(((((((.(((((((((....))))))))).)))))))))))...------..--.----.. ( -29.30) >DroSim_CAF1 2794 97 + 1 GAAAAUCACGAAAAGA--------GCUAUCAACAU---GGCCUAAAGUGGCCAAUGAAUUGGAGUUUAUUUCACGGCGUUGUGAAAUGUGCUCCAACUUUAAC------CC--A----AA ...........((((.--------........(((---((((......)))).)))..(((((((.(((((((((....))))))))).)))))))))))...------..--.----.. ( -29.40) >DroEre_CAF1 1891 107 + 1 AAAAAUCACGCAAAGA--------GCCGUCGACAUAGUGGCCCAAAGUGGCCAAUGAAUUGGAGUUUGUUUCACGGCAUUGUGAAAUAUGCUCCAACUUUAACUUUAACCC-----AAAA ...........((((.--------.............(((((......))))).(((((((((((.(((((((((....))))))))).))))))).)))).)))).....-----.... ( -28.80) >DroYak_CAF1 2953 118 + 1 GAAAAUCACAAAAAGAUAGAAAUCACUAUCAGCAU--UAGUCUAAAGUGGCCAAUGAAAUGGAGUUUAUUUCACAGCGUUGUGAAAUAUGCUCCAACUUUAACUUUGCCCCUCAAAAAAA ..............(((((......))))).(((.--.(((.((((((...........((((((.((((((((......)))))))).))))))))))))))).)))............ ( -29.10) >consensus GAAAAUCACGAAAAGA________GCUAUCAACAU___GGCCUAAAGUGGCCAAUGAAUUGGAGUUUAUUUCACGGCGUUGUGAAAUAUGCUCCAACUUUAAC______CC__A____AA ...........((((.................(((...((((......)))).)))..(((((((.(((((((((....))))))))).))))))))))).................... (-25.46 = -25.38 + -0.08)

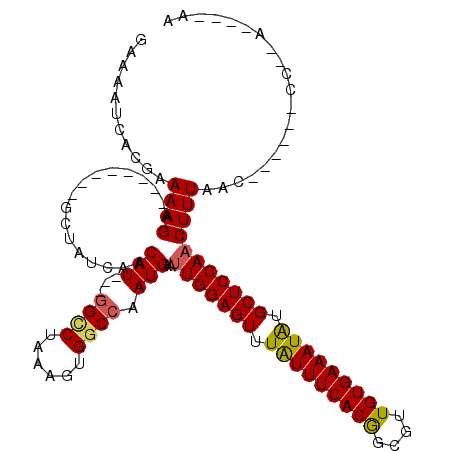
| Location | 2,503,134 – 2,503,231 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.37 |
| Mean single sequence MFE | -29.46 |
| Consensus MFE | -20.30 |
| Energy contribution | -21.26 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.69 |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.988673 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2503134 97 - 22224390 UU----U--GG------GUUAAAGUUGGAGCAUAUUUCACAACGCCGUGAAAUAAACUCCAAUUCAUUGGUCACUUUAGGCC---AUGUUGAUAGC--------UCUUUUCGUGAUUUUC ..----.--((------((((..(((((((..((((((((......))))))))..)))))))(((.(((((......))))---)...)))))))--------)).............. ( -30.00) >DroSec_CAF1 1820 97 - 1 UU----U--GG------GUUAAAGUUGGAGCACAUUUCACAACGACGUGAAAUAAACUCCAAUUCAUUGGCCACUUUGGGCC---AUGUUGAUAGC--------UCUUUUCGUGAUUUUC ..----.--((------((((..(((((((...(((((((......)))))))...)))))))(((.(((((......))))---)...)))))))--------)).............. ( -30.50) >DroSim_CAF1 2794 97 - 1 UU----U--GG------GUUAAAGUUGGAGCACAUUUCACAACGCCGUGAAAUAAACUCCAAUUCAUUGGCCACUUUAGGCC---AUGUUGAUAGC--------UCUUUUCGUGAUUUUC ..----.--((------((((..(((((((...(((((((......)))))))...)))))))(((.(((((......))))---)...)))))))--------)).............. ( -30.60) >DroEre_CAF1 1891 107 - 1 UUUU-----GGGUUAAAGUUAAAGUUGGAGCAUAUUUCACAAUGCCGUGAAACAAACUCCAAUUCAUUGGCCACUUUGGGCCACUAUGUCGACGGC--------UCUUUGCGUGAUUUUU ....-----((((....(((..((((((((....((((((......))))))....))))))))...(((((......))))).......))).))--------)).............. ( -26.40) >DroYak_CAF1 2953 118 - 1 UUUUUUUGAGGGGCAAAGUUAAAGUUGGAGCAUAUUUCACAACGCUGUGAAAUAAACUCCAUUUCAUUGGCCACUUUAGACUA--AUGCUGAUAGUGAUUUCUAUCUUUUUGUGAUUUUC ....((((((((((...........(((((..(((((((((....)))))))))..)))))........))).)))))))...--.....(((((......))))).............. ( -29.81) >consensus UU____U__GG______GUUAAAGUUGGAGCAUAUUUCACAACGCCGUGAAAUAAACUCCAAUUCAUUGGCCACUUUAGGCC___AUGUUGAUAGC________UCUUUUCGUGAUUUUC .................(((((((((((((..((((((((......))))))))..))))))))(((.((((......))))...))))))))........................... (-20.30 = -21.26 + 0.96)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:56:19 2006