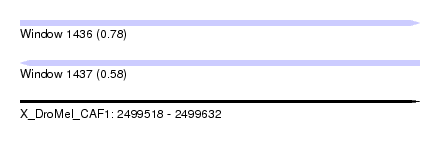
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,499,518 – 2,499,632 |
| Length | 114 |
| Max. P | 0.777307 |

| Location | 2,499,518 – 2,499,632 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 94.47 |
| Mean single sequence MFE | -36.93 |
| Consensus MFE | -30.80 |
| Energy contribution | -30.67 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.777307 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
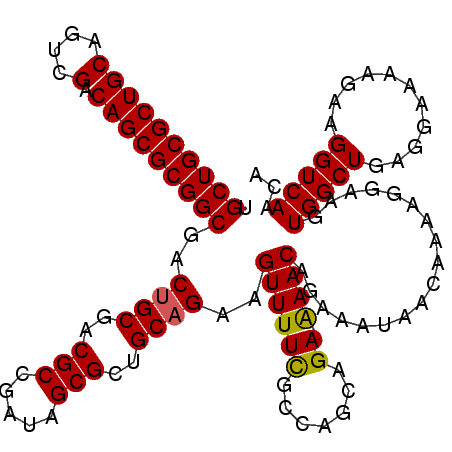
>X_DroMel_CAF1 2499518 114 + 22224390 UGCUGCGCUGCAGUCGACAGCGCGGCGACUGCGACGCCUUUAGCGCUGCAGAAGUUUUCGACAGCAGA-AAACAGAAAUAACAAAAGGAAGUGGCUGAGCAAAAGAAGGUCAACA .((((((((((....).)))))))))..(((((.(((.....))).)))))..((((((.......))-))))..................(((((...(....)..)))))... ( -37.50) >DroSec_CAF1 18127 114 + 1 UGCUGCGCUGCAGUCGACAGCGCGGCGACUGCGACGCCGUUAGCGCUGCAGAAGUUUUCGCCAGCAGA-GAACAGAAAUAACAAAAGGAAGUGGCUGAGGAAAAGAAGGUCAACA .((((((((((....).)))))))))..(((((.(((.....))).)))))..((((((.......))-))))..................(((((...........)))))... ( -36.70) >DroEre_CAF1 18605 114 + 1 UGCUGCGCUGCAGUCGACAGCGCGGCGACGGCUACGCCGAUAGCGCUGCAGAAGUUUUCGCCAGCAGA-AAACAGAAAUAACAAAAGGAAGUGGCUGCGGAAAAGAAGGUCAACA .((((((((((....).)))))))))(((.((((......)))).((((((..((((((.......))-)))).......((........))..))))))........))).... ( -37.60) >DroYak_CAF1 18052 115 + 1 UGCUGCGCUGCUGUCGACAGCGCGGCGACUGCUACGCCGAUAGCGCUGCAGAAGUUUUUGCGAGCAGAGAAACAGAAAUAACAAAAGGAAGUGGCUGAGGAAAAGAAGGUCAACA .((((((((((....).)))))))))((((......((..((((((((((((....))))).)))...............((........)).)))).)).......)))).... ( -35.92) >consensus UGCUGCGCUGCAGUCGACAGCGCGGCGACUGCGACGCCGAUAGCGCUGCAGAAGUUUUCGCCAGCAGA_AAACAGAAAUAACAAAAGGAAGUGGCUGAGGAAAAGAAGGUCAACA .((((((((((....).)))))))))..((((..(((.....)))..))))..((((((.......)).))))..................(((((...........)))))... (-30.80 = -30.67 + -0.12)

| Location | 2,499,518 – 2,499,632 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 94.47 |
| Mean single sequence MFE | -33.38 |
| Consensus MFE | -29.42 |
| Energy contribution | -30.42 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.576317 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
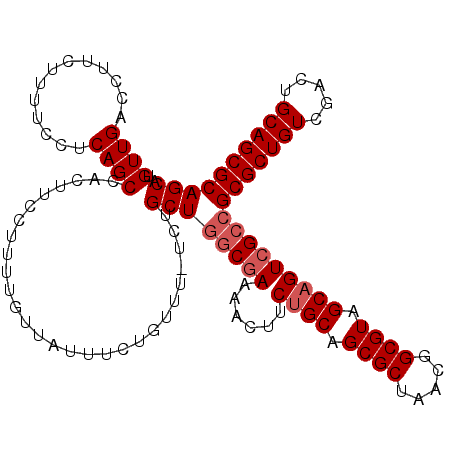
>X_DroMel_CAF1 2499518 114 - 22224390 UGUUGACCUUCUUUUGCUCAGCCACUUCCUUUUGUUAUUUCUGUUU-UCUGCUGUCGAAAACUUCUGCAGCGCUAAAGGCGUCGCAGUCGCCGCGCUGUCGACUGCAGCGCAGCA .(((((.(.......).)))))....................((((-((.......))))))..((((.((((.....)))).))))..((.(((((((.....))))))).)). ( -34.40) >DroSec_CAF1 18127 114 - 1 UGUUGACCUUCUUUUCCUCAGCCACUUCCUUUUGUUAUUUCUGUUC-UCUGCUGGCGAAAACUUCUGCAGCGCUAACGGCGUCGCAGUCGCCGCGCUGUCGACUGCAGCGCAGCA .(((((...........)))))........................-...((((((((......((((.((((.....)))).)))))))))(((((((.....)))))))))). ( -36.70) >DroEre_CAF1 18605 114 - 1 UGUUGACCUUCUUUUCCGCAGCCACUUCCUUUUGUUAUUUCUGUUU-UCUGCUGGCGAAAACUUCUGCAGCGCUAUCGGCGUAGCCGUCGCCGCGCUGUCGACUGCAGCGCAGCA .((((......((((((.((((........................-...))))).)))))......))))(((...((((.......))))(((((((.....)))))))))). ( -31.93) >DroYak_CAF1 18052 115 - 1 UGUUGACCUUCUUUUCCUCAGCCACUUCCUUUUGUUAUUUCUGUUUCUCUGCUCGCAAAAACUUCUGCAGCGCUAUCGGCGUAGCAGUCGCCGCGCUGUCGACAGCAGCGCAGCA .(((((...........)))))..........................(((((.(((........))).((((.....)))))))))..((.(((((((.....))))))).)). ( -30.50) >consensus UGUUGACCUUCUUUUCCUCAGCCACUUCCUUUUGUUAUUUCUGUUU_UCUGCUGGCGAAAACUUCUGCAGCGCUAACGGCGUAGCAGUCGCCGCGCUGUCGACUGCAGCGCAGCA .((((.............))))............................((((((((......((((.((((.....)))).)))))))))(((((((.....)))))))))). (-29.42 = -30.42 + 1.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:56:15 2006