| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,477,505 – 2,477,607 |
| Length | 102 |
| Max. P | 0.748082 |

| Location | 2,477,505 – 2,477,607 |
|---|---|
| Length | 102 |
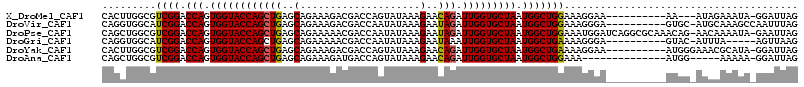
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 80.62 |
| Mean single sequence MFE | -25.02 |
| Consensus MFE | -19.67 |
| Energy contribution | -19.37 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.748082 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
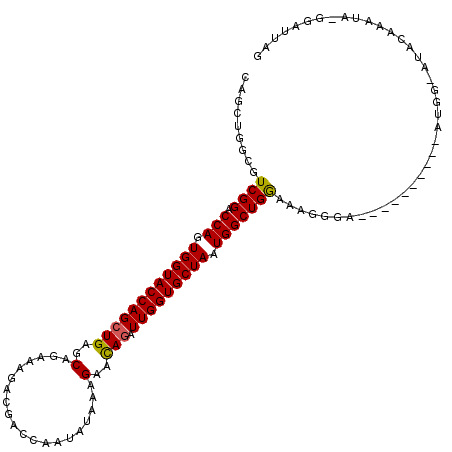
>X_DroMel_CAF1 2477505 102 - 22224390 CACUUGGCGUCGGACCAGUGGUACCAGCUGAGCAGAAAGACGACCAGUAUAAAGAACAGAUUGGUGCUAAUGGCUGGAAAGGAA----------AA---AUAGAAAUA-GGAUUAG ..(((....((((.(((.((((((((((((..(......((.....)).....)..))).))))))))).))))))).)))...----------..---.........-....... ( -24.30) >DroVir_CAF1 2608 105 - 1 CAGGUGGCAUCGGACCAGUGGUACCAGCUGAGCAGAAAGACGACCAAUAUAAAGAAUAGAUUGGUGCUAAUGGCUGGAAAGGGA----------GUGC-AUGCAAAGCCAAUUUAG ..(((.((((((..((..(....((((((.(((.(.....).((((((.((.....)).)))))))))...)))))).)..)).----------.)).-))))...)))....... ( -26.80) >DroPse_CAF1 8776 114 - 1 CAGCUGGCGUCGGACCAGUGGUACCAGCUGAGCAGAAAAACGACCAAUAUAAAGAAUAGAUUGGUGCUAAUGGCUGGAAAUGGAUCAGGCGCAAACAG-AACAAAAUA-GAAUUAG ...((((((((.(((((......((((((.(((.(.....).((((((.((.....)).)))))))))...))))))...))).)).)))))...)))-.........-....... ( -28.90) >DroGri_CAF1 4450 100 - 1 CAGGUGGCAUCGGACCAGUGGUACCAGCUGAGCAGAAAAACGACCAAUAUAAAGAAUAAAUUGGUGCUAAUGGCUGAAAAGGGA----------GUAC-AUUUA-----AGUUAAG ....((((.((((.(((.((((((((((...))..............(((.....)))...)))))))).)))))))...(...----------...)-.....-----.)))).. ( -18.10) >DroYak_CAF1 2175 105 - 1 CACUUGGCGUCGGACCAGUGGUACCAGCUGAGCAGAAAGACGACCAGUAUAAAGAACAGAUUGGUGCUAAUGGCUGAAAAGGAA----------AUGGGAAACGCAUA-GGAUUAG ..((((((.((((.(((.((((((((((((..(......((.....)).....)..))).))))))))).))))))).......----------...(....))).))-))..... ( -29.30) >DroAna_CAF1 3508 96 - 1 CAGCUGGCGUCGGACCAGUGGUACCAGCUGAGCAGAAAGAUGACCAGUAUAAAGAACAGAUUGGUGCUAAUGGCUGGAAA--------------AUGG-----AAAAA-GGAUUAG (((((((..(((......)))..))))))).............(((((....((.((......)).))....)))))...--------------....-----.....-....... ( -22.70) >consensus CAGCUGGCGUCGGACCAGUGGUACCAGCUGAGCAGAAAGACGACCAAUAUAAAGAACAGAUUGGUGCUAAUGGCUGGAAAGGGA__________AUGG_AUACAAAUA_GGAUUAG .........((((.(((.((((((((((((..(....................)..))).))))))))).)))))))....................................... (-19.67 = -19.37 + -0.31)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:56:09 2006