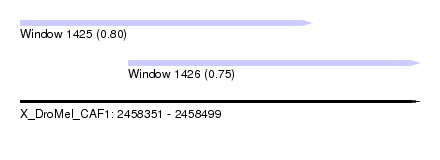
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,458,351 – 2,458,499 |
| Length | 148 |
| Max. P | 0.800404 |

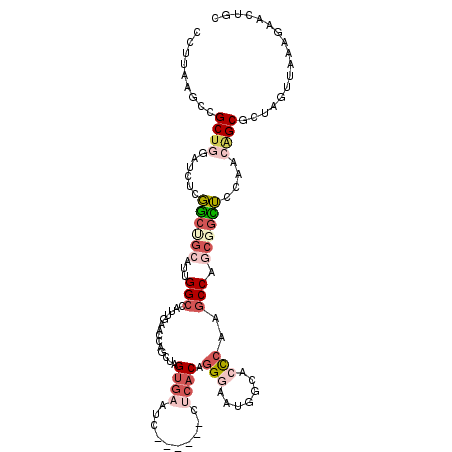
| Location | 2,458,351 – 2,458,459 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 74.91 |
| Mean single sequence MFE | -36.90 |
| Consensus MFE | -16.80 |
| Energy contribution | -18.00 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.800404 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2458351 108 + 22224390 UGUAAUUUCCUAUUCCUGUCCUGCAGGGUGCGCACCUGUUCCUUAAGCCGCUGGAUCUCGGCUGCAUUGGCCAUUGAACCAGCUAGUGACUCCUC------ACAGGGAAUGGCA .........((((((((.....(((((.......)))))..........(((((.((..((((.....))))...)).)))))..((((....))------)).)))))))).. ( -34.60) >DroPse_CAF1 5644 111 + 1 UGUAGCUUACGGGUCUUCUCCUGCAGCGUCUUUACCAAAUCCUUUAGACCCUGUUGGUCAGCAGCA---GCCGUAGAGGAUGCUGAUGUACCAUCAGCGCCACUUGGCAGCGGA ....(((..((((......)))).))).......((..((((((((....((((((.....)))))---)...))))))))(((((((...)))))))(((....)))...)). ( -37.40) >DroSec_CAF1 2421 108 + 1 UGUAAUUUCCUAUUCCUGUCCUGCAGGGUGCGCACCUGAUUCUUAAGCAGCUGGAUCUCGGCCGCAUUGGCCAUUGAACCAGCUAGUGAAUCCUC------ACAGGAAAUGGCA .((.(((((((...((((.....))))(((.(.....(((((....((((((((.((..((((.....))))...)).)))))).)))))))).)------))))))))).)). ( -38.30) >DroSim_CAF1 3249 108 + 1 UGUAAUUUCCUAUUCCUGUCCUGCAGGCUACGCACCUGAUCCUUAAGCAGCUGGAUCUCGGCCGCAUUGGCCAUUGAACCAGCUAGUGAAUCCUC------ACAGGAAAUGGCA .((.(((((((...((((.....)))).....................((((((.((..((((.....))))...)).)))))).((((....))------))))))))).)). ( -35.50) >DroEre_CAF1 17894 108 + 1 UGUAGUUUUUUAAUCCUGUCCUGCAGGGUGCGCACUUCUUCCUUGUGGCGCUUAUACUCGGCUUCAUUGGCUACUGAACCAGCUAGUGAAUCCCC------ACAGGGAAUCUCA .(.(((......((((((.....))))))....))).)((((((((((.(((.......)))((((((((((........))))))))))...))------))))))))..... ( -35.00) >DroYak_CAF1 16302 108 + 1 UGCAGUUUCCUGUUCCUGUCCAGCAGGGUGCGCACCUGUUCCUUGACGCGCUGGUACUCGGCUGCAUUGGCCAUUGAACCAGCUAGUGACUCCUC------ACAGGGAAUAGCA (((.((.(((((((.......))))))).))))).(((((((((.....((((((.(..((((.....))))...).))))))..((((....))------))))))))))).. ( -40.60) >consensus UGUAAUUUCCUAUUCCUGUCCUGCAGGGUGCGCACCUGAUCCUUAAGCCGCUGGAUCUCGGCUGCAUUGGCCAUUGAACCAGCUAGUGAAUCCUC______ACAGGGAAUGGCA .((.(((((((..(((((.....))))).....................(((((.....((((.....))))......)))))....................))))))).)). (-16.80 = -18.00 + 1.20)

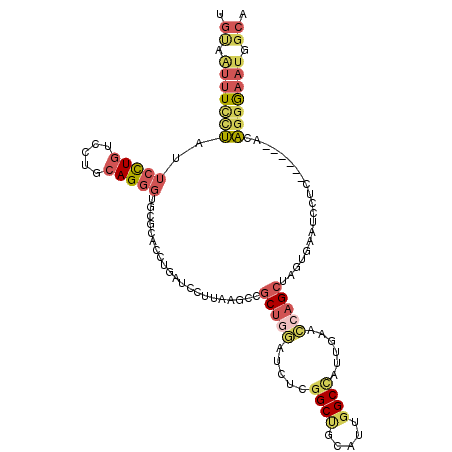
| Location | 2,458,391 – 2,458,499 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 74.91 |
| Mean single sequence MFE | -38.20 |
| Consensus MFE | -14.14 |
| Energy contribution | -15.95 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.37 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.752788 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2458391 108 + 22224390 CCUUAAGCCGCUGGAUCUCGGCUGCAUUGGCCAUUGAACCAGCUAGUGACUC------CUCACAGGGAAUGGCACCCAAGCCUGCGGCUCCAACAGCGCUAAUUAAGGAACUGC ((((((((((((((.((..((((.....))))...)).)))))..((((...------.)))).......))).....((((((.(....)..))).)))..))))))...... ( -35.20) >DroSec_CAF1 2461 108 + 1 UCUUAAGCAGCUGGAUCUCGGCCGCAUUGGCCAUUGAACCAGCUAGUGAAUC------CUCACAGGAAAUGGCACCCAAGCCUACGGCUCCAACAGCGCUGGUUAAGGAACUGC ((((((.(((((((.((..((((.....))))...)).)))(((.((((...------.)))).(((...(((......)))......)))...))))))).))))))...... ( -36.80) >DroSim_CAF1 3289 107 + 1 CCUUAAGCAGCUGGAUCUCGGCCGCAUUGGCCAUUGAACCAGCUAGUGAAUC------CUCACAGGAAAUGGCACCCAAGCCUACGGCUCCAACAGCGCUGGUUAA-GAACUAC .(((((.(((((((.((..((((.....))))...)).)))(((.((((...------.)))).(((...(((......)))......)))...))))))).))))-)...... ( -36.40) >DroEre_CAF1 17934 108 + 1 CCUUGUGGCGCUUAUACUCGGCUUCAUUGGCUACUGAACCAGCUAGUGAAUC------CCCACAGGGAAUCUCACCCAAGCCAGCUAUUUCAACAGCUCUACUUAUAGAGCUGG ((((((((.(((.......)))((((((((((........))))))))))..------.))))))))..........................((((((((....)))))))). ( -36.80) >DroYak_CAF1 16342 108 + 1 CCUUGACGCGCUGGUACUCGGCUGCAUUGGCCAUUGAACCAGCUAGUGACUC------CUCACAGGGAAUAGCACCCAAGCCAGCGGCUCCAACUGCACUAGUUAUAGAGCUGC .......((((((((.(..((((.....))))...).))))))..((((...------.)))).(((.......)))..))..(((((((.(((((...)))))...))))))) ( -35.70) >DroAna_CAF1 17254 114 + 1 CCUUCAGCCGCUUGAUCUCCUCGGCCUGGGCAGCGGAUCCUGAAAGGGAAUCGGCCUCCGAACUCGGCACAGAACUCAACCCAGCAGAGCCCAUGGCACUGGUGAUCGAGCUGC ......((.((((((((.((...((((((((.((.((((((....))).))).))..(((....))).....................))))).)))...)).)))))))).)) ( -48.30) >consensus CCUUAAGCCGCUGGAUCUCGGCUGCAUUGGCCAUUGAACCAGCUAGUGAAUC______CUCACAGGGAAUGGCACCCAAGCCAGCGGCUCCAACAGCGCUAGUUAAAGAACUGC .........((((......((((((...(((..............((((..........)))).(((.......)))..))).))))))....))))................. (-14.14 = -15.95 + 1.81)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:56:05 2006