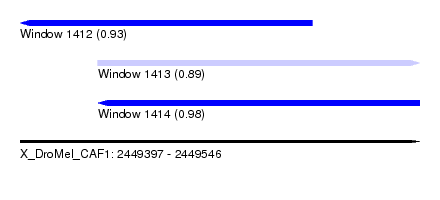
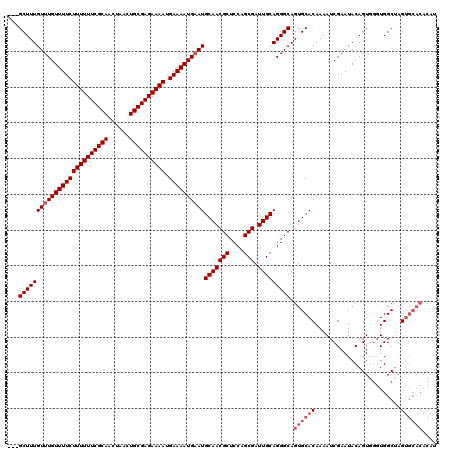
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,449,397 – 2,449,546 |
| Length | 149 |
| Max. P | 0.975629 |

| Location | 2,449,397 – 2,449,506 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.92 |
| Mean single sequence MFE | -40.30 |
| Consensus MFE | -31.59 |
| Energy contribution | -33.59 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.73 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928909 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2449397 109 - 22224390 CUCGCUUUGUUUGUUUUCUUUUUUCGCAACUAACUGCGAGAAAAUGAAAAUGCAUGCAACGCUCCAGCGAUUGCAGGGCAGUGCACAAAAUCGAAUACAGUU--UGGCGAG--------- ((((((....((((.(((((((((((((......))))))))))((....(((.((((((((....))).)))))..)))...)).......))).))))..--.))))))--------- ( -37.80) >DroSec_CAF1 9965 117 - 1 ---GCUUUGUUUGUUUUCUUUUUUCGCAACUAACUGCGAGAAAAUGAAAAUGAAUGCAACGCUCCAGCGAUUGCAGGGCAGUGCACAAAAUCGAAUACAGUGGGUGGCUAGUGCACACAU ---((((((((..(((((((((((((((......)))))))))).)))))..)))(((((((....))).))))))))).((((((...(((.(......).))).....)))))).... ( -41.00) >DroSim_CAF1 12354 117 - 1 ---GCUUUGUUUGUUUUCUUUUUUCGCAACUAACUGCGAGAAAAUGAAAAUGAAUGCAACGCUCCAGCGAUUGCAGGGCAGUGCACAAAAUCGAAUACAGUGGGUGGCUGGUGCACACAU ---((((((((..(((((((((((((((......)))))))))).)))))..)))(((((((....))).))))))))).((((((...(((.(......).))).....)))))).... ( -42.10) >consensus ___GCUUUGUUUGUUUUCUUUUUUCGCAACUAACUGCGAGAAAAUGAAAAUGAAUGCAACGCUCCAGCGAUUGCAGGGCAGUGCACAAAAUCGAAUACAGUGGGUGGCUAGUGCACACAU ...(((((((((((((((((((((((((......)))))))))).))))))))))(((((((....))).))))))))).((((((........................)))))).... (-31.59 = -33.59 + 2.00)

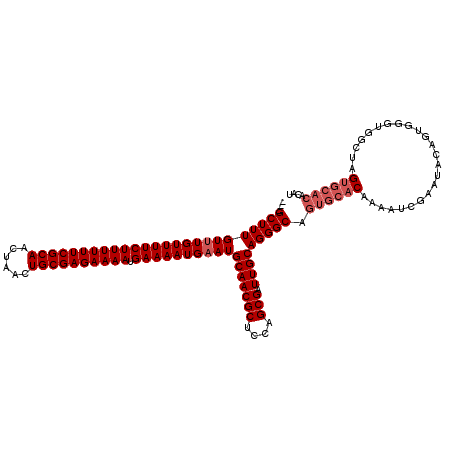
| Location | 2,449,426 – 2,449,546 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.63 |
| Mean single sequence MFE | -32.30 |
| Consensus MFE | -29.79 |
| Energy contribution | -29.57 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.888684 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2449426 120 + 22224390 UGCCCUGCAAUCGCUGGAGCGUUGCAUGCAUUUUCAUUUUCUCGCAGUUAGUUGCGAAAAAAGAAAACAAACAAAGCGAGCGAAUGAGAGGCGCAAUAUAUAGUAUAGUAAAGUGCUAUU (((..(((((.(((....)))))))).((.(((((((((.(((((.(((.(((............))).)))...))))).))))))))))))))....(((((((......))))))). ( -35.60) >DroSec_CAF1 10005 116 + 1 UGCCCUGCAAUCGCUGGAGCGUUGCAUUCAUUUUCAUUUUCUCGCAGUUAGUUGCGAAAAAAGAAAACAAACAAAGC----GAAUGAGAGGCGCAAUAUAUAGUAUAGUAAAGUGCUAUU ((((((.((.((((((((((...)).))).(((((.((((.((((((....)))))).))))))))).......)))----)).))..))).)))....(((((((......))))))). ( -30.90) >DroSim_CAF1 12394 116 + 1 UGCCCUGCAAUCGCUGGAGCGUUGCAUUCAUUUUCAUUUUCUCGCAGUUAGUUGCGAAAAAAGAAAACAAACAAAGC----GAAUGAGAGGCGCGAUAUAUAGUAUAGUAAAGUGCUAUU .(((((.((.((((((((((...)).))).(((((.((((.((((((....)))))).))))))))).......)))----)).)))).))).......(((((((......))))))). ( -30.40) >consensus UGCCCUGCAAUCGCUGGAGCGUUGCAUUCAUUUUCAUUUUCUCGCAGUUAGUUGCGAAAAAAGAAAACAAACAAAGC____GAAUGAGAGGCGCAAUAUAUAGUAUAGUAAAGUGCUAUU ((((((((((.(((....))))))).....(((((.((((.((((((....)))))).))))))))).....................))).)))....(((((((......))))))). (-29.79 = -29.57 + -0.22)

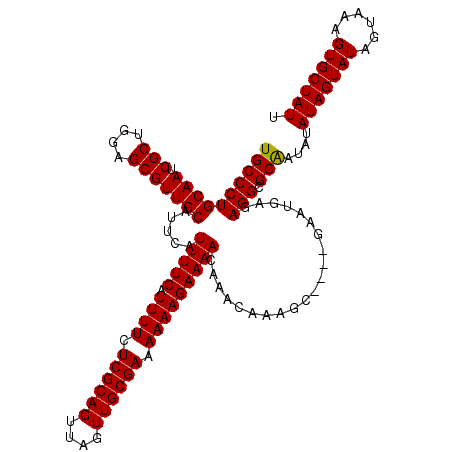
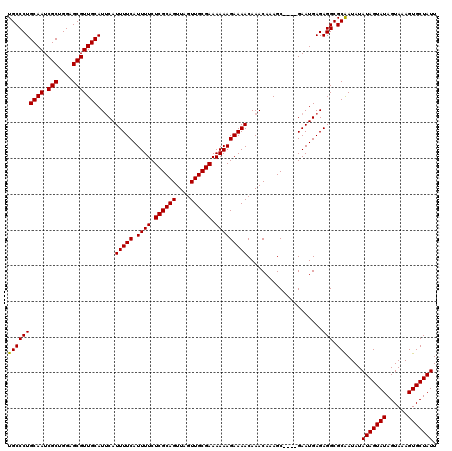
| Location | 2,449,426 – 2,449,546 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.63 |
| Mean single sequence MFE | -34.77 |
| Consensus MFE | -32.13 |
| Energy contribution | -32.47 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -4.03 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.975629 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2449426 120 - 22224390 AAUAGCACUUUACUAUACUAUAUAUUGCGCCUCUCAUUCGCUCGCUUUGUUUGUUUUCUUUUUUCGCAACUAACUGCGAGAAAAUGAAAAUGCAUGCAACGCUCCAGCGAUUGCAGGGCA ....(((...((........))...)))((((..((.(((((.((.((((.(((((((((((((((((......)))))))))).))))..))).)))).))...))))).))..)))). ( -37.00) >DroSec_CAF1 10005 116 - 1 AAUAGCACUUUACUAUACUAUAUAUUGCGCCUCUCAUUC----GCUUUGUUUGUUUUCUUUUUUCGCAACUAACUGCGAGAAAAUGAAAAUGAAUGCAACGCUCCAGCGAUUGCAGGGCA ....(((...((........))...)))((((..((.((----(((..(((..(((((((((((((((......)))))))))).)))))..)))((...))...))))).))..)))). ( -33.80) >DroSim_CAF1 12394 116 - 1 AAUAGCACUUUACUAUACUAUAUAUCGCGCCUCUCAUUC----GCUUUGUUUGUUUUCUUUUUUCGCAACUAACUGCGAGAAAAUGAAAAUGAAUGCAACGCUCCAGCGAUUGCAGGGCA .((((.......))))............((((..((.((----(((..(((..(((((((((((((((......)))))))))).)))))..)))((...))...))))).))..)))). ( -33.50) >consensus AAUAGCACUUUACUAUACUAUAUAUUGCGCCUCUCAUUC____GCUUUGUUUGUUUUCUUUUUUCGCAACUAACUGCGAGAAAAUGAAAAUGAAUGCAACGCUCCAGCGAUUGCAGGGCA .((((.......))))............((((................((((((((((((((((((((......)))))))))).))))))))))(((((((....))).)))).)))). (-32.13 = -32.47 + 0.33)

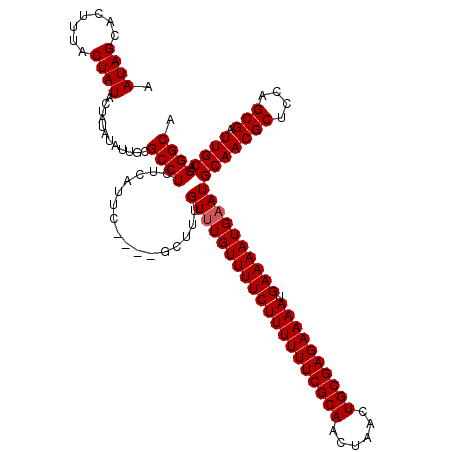
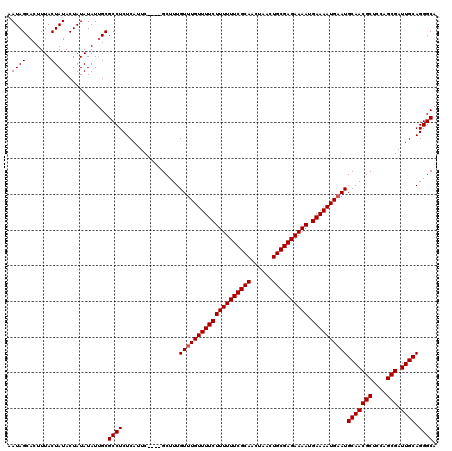
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:55:54 2006