
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 22,074,146 – 22,074,343 |
| Length | 197 |
| Max. P | 0.895769 |

| Location | 22,074,146 – 22,074,263 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.06 |
| Mean single sequence MFE | -35.51 |
| Consensus MFE | -28.91 |
| Energy contribution | -29.14 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.81 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.518886 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 22074146 117 - 22224390 AUUGGACAUAAUCCGGGCCAUUUGAGAGUAAUCCCGGAGUGAACCGGGAUAAUUCAAACGGCCAGAUCGCUCAACAGGAUUCCCAAAGGG---GGCCACUCAAAAUCAUCACCCUAUCGC .((((....(((((.((((.((((((....(((((((......)))))))..)))))).)))).((....))....))))).)))).(((---(.................))))..... ( -36.03) >DroSec_CAF1 139423 120 - 1 AUUAGACAUAACCCGGGCCAUUUAAGAGUAAUUCCGGAGCGAUCUGGGAUAAUCCAAACGGCCAGAUCGCUCAACAGGAUUCCCAAAGGGUCGGGCCACCCAAAAUCAUCACCCUAUCGC ...........(((((.((.(((..(((....(((.(((((((((((......((....)))))))))))))....))))))..))))).)))))......................... ( -35.40) >DroSim_CAF1 111902 120 - 1 AUUAGACAUAACCCGGGCCAUUUAAGAGUAAUUCCGGAGCGAUCUGGGAUAAUCCAAACGGCCAGAUCGCUCAACAGGAUUCCCAAAGGGGCGGGCCACCCAAAAUCAUCACCCUAUCGC ...............((((.............(((.(((((((((((......((....)))))))))))))....)))..(((....)))..))))....................... ( -35.10) >consensus AUUAGACAUAACCCGGGCCAUUUAAGAGUAAUUCCGGAGCGAUCUGGGAUAAUCCAAACGGCCAGAUCGCUCAACAGGAUUCCCAAAGGG_CGGGCCACCCAAAAUCAUCACCCUAUCGC ...............((((..........(((((..(((((((((((..............)))))))))))....)))))(((...)))...))))....................... (-28.91 = -29.14 + 0.23)


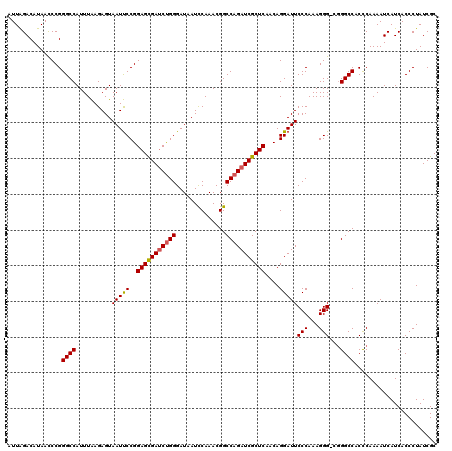
| Location | 22,074,183 – 22,074,303 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.67 |
| Mean single sequence MFE | -45.77 |
| Consensus MFE | -36.26 |
| Energy contribution | -36.27 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.16 |
| Mean z-score | -3.39 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.895769 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 22074183 120 + 22224390 AUCCUGUUGAGCGAUCUGGCCGUUUGAAUUAUCCCGGUUCACUCCGGGAUUACUCUCAAAUGGCCCGGAUUAUGUCCAAUGACCCAAAUCGGGAGGGGAUUAGAAAUAUGACGAAAUAGG ..((((((..(((((((((((((((((...(((((((......))))))).....)))))))).)))))))..((((.....(((.....)))...)))).........).)..)))))) ( -44.70) >DroSec_CAF1 139463 120 + 1 AUCCUGUUGAGCGAUCUGGCCGUUUGGAUUAUCCCAGAUCGCUCCGGAAUUACUCUUAAAUGGCCCGGGUUAUGUCUAAUAGCCCAAGUCGGGAGAGGAUUAGGAAUAUGGCGCAAUAGG ..(((((((.(((((((((((....))......)))))))))(((..((((.(((((...((((..(((((((.....)))))))..)))).))))))))).)))........))))))) ( -46.30) >DroSim_CAF1 111942 120 + 1 AUCCUGUUGAGCGAUCUGGCCGUUUGGAUUAUCCCAGAUCGCUCCGGAAUUACUCUUAAAUGGCCCGGGUUAUGUCUAAUAGCCCAAGUCGGGAGAGGAUUAGGAAUAUGGCGCAAUAGG ..(((((((.(((((((((((....))......)))))))))(((..((((.(((((...((((..(((((((.....)))))))..)))).))))))))).)))........))))))) ( -46.30) >consensus AUCCUGUUGAGCGAUCUGGCCGUUUGGAUUAUCCCAGAUCGCUCCGGAAUUACUCUUAAAUGGCCCGGGUUAUGUCUAAUAGCCCAAGUCGGGAGAGGAUUAGGAAUAUGGCGCAAUAGG ..(((((((((((((((((..............)))))))))))...((((.(((((...((((..(((((((.....)))))))..)))).))))))))).............)))))) (-36.26 = -36.27 + 0.01)

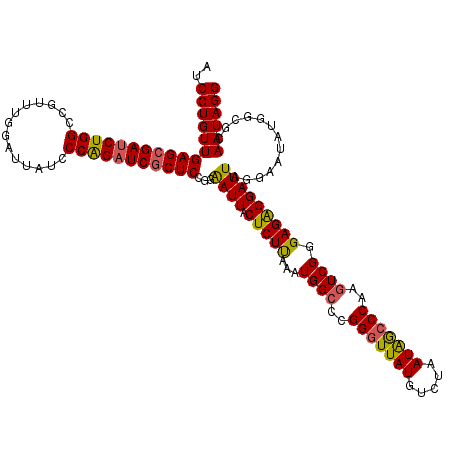
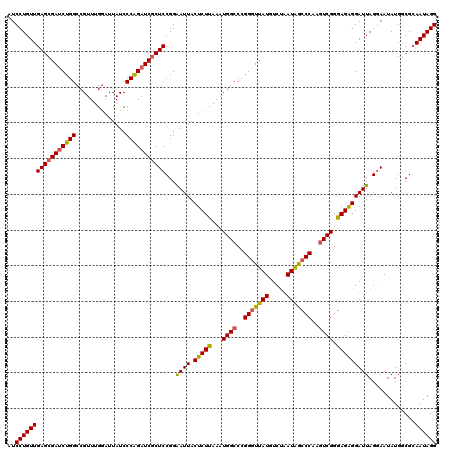
| Location | 22,074,223 – 22,074,343 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.33 |
| Mean single sequence MFE | -34.93 |
| Consensus MFE | -29.67 |
| Energy contribution | -29.57 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.85 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.517833 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 22074223 120 + 22224390 ACUCCGGGAUUACUCUCAAAUGGCCCGGAUUAUGUCCAAUGACCCAAAUCGGGAGGGGAUUAGAAAUAUGACGAAAUAGGGCCCCCGGCAAUAUUGCAUUGGAAGACAGGAAAAUACAUA ..((((((....(........).)))))).....(((((((.(((.....))).((((..(.....(((......))).)..))))..........)))))))................. ( -30.00) >DroSec_CAF1 139503 116 + 1 GCUCCGGAAUUACUCUUAAAUGGCCCGGGUUAUGUCUAAUAGCCCAAGUCGGGAGAGGAUUAGGAAUAUGGCGCAAUAGGGCGCCGGGCAAUAUUGCAUUGGAAGUCAGGAAAAU----A ..(((.......(((((...((((..(((((((.....)))))))..)))).)))))((((.......((((((......)))))).(((....)))......)))).)))....----. ( -37.40) >DroSim_CAF1 111982 116 + 1 GCUCCGGAAUUACUCUUAAAUGGCCCGGGUUAUGUCUAAUAGCCCAAGUCGGGAGAGGAUUAGGAAUAUGGCGCAAUAGGGCGCCGGGCAAUAUUGCAUUGGAAGUCAGGAAAAU----A ..(((.......(((((...((((..(((((((.....)))))))..)))).)))))((((.......((((((......)))))).(((....)))......)))).)))....----. ( -37.40) >consensus GCUCCGGAAUUACUCUUAAAUGGCCCGGGUUAUGUCUAAUAGCCCAAGUCGGGAGAGGAUUAGGAAUAUGGCGCAAUAGGGCGCCGGGCAAUAUUGCAUUGGAAGUCAGGAAAAU____A ..(((((((((.(((((...((((..(((((((.....)))))))..)))).)))))))))...........((((((..((.....))..)))))).)))))................. (-29.67 = -29.57 + -0.10)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:10:28 2006