
| Sequence ID | X_DroMel_CAF1 |
|---|---|
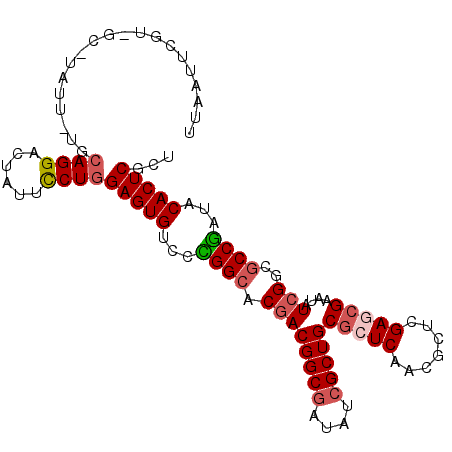
| Location | 22,071,117 – 22,071,219 |
| Length | 102 |
| Max. P | 0.818908 |

| Location | 22,071,117 – 22,071,219 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 79.90 |
| Mean single sequence MFE | -36.82 |
| Consensus MFE | -27.19 |
| Energy contribution | -27.75 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.818908 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 22071117 102 + 22224390 UUAAUUCGU-UC-CAUU-UUCAGGACUAUUCCUGGAGUGCCCCGGCACGACGGCGAUAUCGCUGCGCUCAACACUCGAGAGAAUAUCGGCGCCGAUCCACUCGCU ......(((-(.-....-(((((((....)))))))((((....))))))))((((.((((..(((((......((....)).....)))))))))....)))). ( -32.90) >DroVir_CAF1 126103 94 + 1 ----------GU-UAUUUCGCAGGCCUGUUUCUGGAGUGUCCUGGCACGACGGCGCUGUCGCUGCGCUCGACGCUGGAACGCAUCUCGGCGCCAAUACACUCGCU ----------..-.......((((......))))((((((..((((....((((((.......)))).))..((((((.....)).))))))))..))))))... ( -32.90) >DroGri_CAF1 92211 102 + 1 --UAUCGGUUGCAUGUU-CGCAGGACUUUUCCUCGAGUGUCCUGGCACUACGGCUCUGUCGCUGCGCUCAACGCUGGAACGCAUCUCGGCGCCGAUACACUCGUU --(((((((....((..-((((((((....((...(((((....)))))..))....))).)))))..))..((((((.....)).)))))))))))........ ( -35.10) >DroSim_CAF1 107733 102 + 1 UUAAUUCGC-CC-UAUU-UUCAGGACUUUUCCUGGAGUGUCCCGGCACGACGGCGAUAUCGCUGCGCUCAACCCUCGAGCGAAUAUCGGCGCCGAUCCACUCGCU .......((-..-....-..(((((....)))))(((((...((((.((((((((....)))))(((((.......)))))....)))..))))...))))))). ( -38.70) >DroYak_CAF1 113072 102 + 1 UGAAAUUGU-CC-GAUU-UUCAGGACUAUUCCUGGAGUGUCCUGGCACGACGGCGAUAUCGCUGCGCUCAACCCUCGAGCGAAUAUCGGCGCCGAUCCACUCGCU .((((((..-..-))))-))(((((....)))))(((((...((((.((((((((....)))))(((((.......)))))....)))..))))...)))))... ( -36.50) >DroMoj_CAF1 103668 103 + 1 UUGAGCUGUGGC-UAUU-UGCAGGCCUCUUUCUGGAGUGUCCCGGGACGACGGCGCUGUCGCUGCGCUCGACGCUGGAGCGCAUCUCGGCGCCGAUACACUCGCU ..(((.((((((-(...-....))))((.((((((......)))))).))((((((((....(((((((.......)))))))...))))))))..))))))... ( -44.80) >consensus UUAAUUCGU_GC_UAUU_UGCAGGACUAUUCCUGGAGUGUCCCGGCACGACGGCGAUAUCGCUGCGCUCAACGCUCGAGCGAAUAUCGGCGCCGAUACACUCGCU ....................((((......))))(((((...((((.((((((((....)))))(((((.......)))))....)))..))))...)))))... (-27.19 = -27.75 + 0.56)


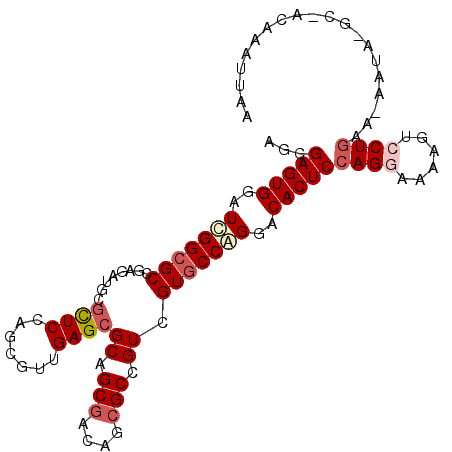
| Location | 22,071,117 – 22,071,219 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 79.90 |
| Mean single sequence MFE | -37.95 |
| Consensus MFE | -26.56 |
| Energy contribution | -27.50 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.802753 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 22071117 102 - 22224390 AGCGAGUGGAUCGGCGCCGAUAUUCUCUCGAGUGUUGAGCGCAGCGAUAUCGCCGUCGUGCCGGGGCACUCCAGGAAUAGUCCUGAA-AAUG-GA-ACGAAUUAA .((((....((((((((((((((((....)))))))).))))..)))).))))((((((((....))))..(((((....)))))..-....-).-)))...... ( -38.00) >DroVir_CAF1 126103 94 - 1 AGCGAGUGUAUUGGCGCCGAGAUGCGUUCCAGCGUCGAGCGCAGCGACAGCGCCGUCGUGCCAGGACACUCCAGAAACAGGCCUGCGAAAUA-AC---------- .((((((((.(((((((.(((((((......)))))(.((((.......))))).))))))))).)))))).((........))))......-..---------- ( -35.10) >DroGri_CAF1 92211 102 - 1 AACGAGUGUAUCGGCGCCGAGAUGCGUUCCAGCGUUGAGCGCAGCGACAGAGCCGUAGUGCCAGGACACUCGAGGAAAAGUCCUGCG-AACAUGCAACCGAUA-- ..(((((((...(((((....(((.((((...(((((....)))))...))))))).)))))...)))))))((((....))))((.-.....))........-- ( -35.20) >DroSim_CAF1 107733 102 - 1 AGCGAGUGGAUCGGCGCCGAUAUUCGCUCGAGGGUUGAGCGCAGCGAUAUCGCCGUCGUGCCGGGACACUCCAGGAAAAGUCCUGAA-AAUA-GG-GCGAAUUAA .(((((((..(((((((.(((...(((((((...)))))))..(((....))).))))))))))..)))))(((((....)))))..-....-..-))....... ( -42.80) >DroYak_CAF1 113072 102 - 1 AGCGAGUGGAUCGGCGCCGAUAUUCGCUCGAGGGUUGAGCGCAGCGAUAUCGCCGUCGUGCCAGGACACUCCAGGAAUAGUCCUGAA-AAUC-GG-ACAAUUUCA .(.(((((..(.(((((.(((...(((((((...)))))))..(((....))).)))))))).)..)))))).(((((.((((....-....-))-)).))))). ( -39.00) >DroMoj_CAF1 103668 103 - 1 AGCGAGUGUAUCGGCGCCGAGAUGCGCUCCAGCGUCGAGCGCAGCGACAGCGCCGUCGUCCCGGGACACUCCAGAAAGAGGCCUGCA-AAUA-GCCACAGCUCAA (((((((((..((((((...(((((......)))))..)))).(((((......)))))..))..))))))........(((.....-....-)))...)))... ( -37.60) >consensus AGCGAGUGGAUCGGCGCCGAGAUGCGCUCCAGCGUUGAGCGCAGCGACAGCGCCGUCGUGCCAGGACACUCCAGGAAAAGUCCUGAA_AAUA_GC_ACAAAUUAA ...(((((..(((((((........((((.......))))((.(((....))).)).)))))))..)))))((((......)))).................... (-26.56 = -27.50 + 0.94)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:10:25 2006