
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 22,069,019 – 22,069,164 |
| Length | 145 |
| Max. P | 0.861075 |

| Location | 22,069,019 – 22,069,135 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.40 |
| Mean single sequence MFE | -36.76 |
| Consensus MFE | -26.60 |
| Energy contribution | -26.32 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.861075 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
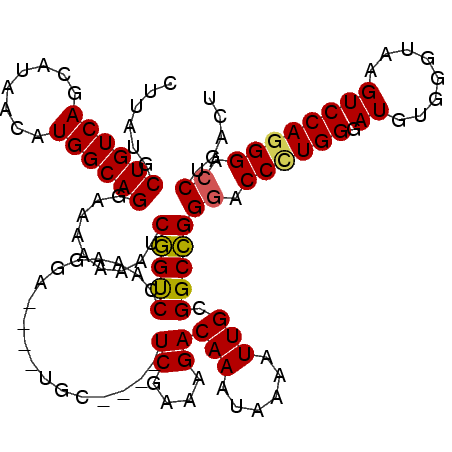
>X_DroMel_CAF1 22069019 116 + 22224390 CUUAUGCUGUCAGCAUAACAUGGCAGGAAAAAAAUCGGUCCAAAGGCUGGCUGC----UCGAAAGACAAAUAAAAUUGCGGCCGGGACCCUGGAAUGUGGGUAAGUCCAGGGACCUGACU ....(.((((((........)))))).)........(((((...((((((((((----((....))...........)))))))).((((........))))....))..)))))..... ( -39.00) >DroSec_CAF1 127002 111 + 1 CUUAUGCUGUCAGCAUAACAUGGCAGGAAAAAAAUCGGCCCAAAGGA----UGC----UCGAAAGACAAAUAAAAUUGCGGCCGGGACCCUGGUAUGUGGGUAAGUCCAGGGACCUGAC- .((((((.....)))))).....((((.......(((((((((...(----(..----((....))...))....))).))))))..((((((.((........)))))))).))))..- ( -36.20) >DroSim_CAF1 105774 111 + 1 CUUAUGCUGUCAGCAUAACAUGGCAGGAAAAAAAUCGGCCCAAAGGA----UGC----UCGAAAGACAAAUAAAAUUGCGGCCGGGACCCUGGUAUGUGGGUAAGUCCAGGGACCUGAC- .((((((.....)))))).....((((.......(((((((((...(----(..----((....))...))....))).))))))..((((((.((........)))))))).))))..- ( -36.20) >DroEre_CAF1 101442 116 + 1 CUUAUGCUGUCAGCAUAACAUGGCAGAAAAAAGUUCGGUCCAAAGGA----CGCUCGCUCGAAAGACAAAUAAAAUUGCGGCCGGGACCGUGGGAUGUGGGCUAGUCCAGGGACCUGACU ......((((((........)))))).....((((.(((((...(((----(((((((((....)).........(..(((......)))..)...))))))..))))..))))).)))) ( -42.00) >DroYak_CAF1 110520 111 + 1 CUUAUGCUGUCAG-AUAACAUGGCAGAAAAAAGCUCAGUCCAAAGGA----CGC----UCAAAAGACAAAUAAAAUUGUGGCUGGGACCUUGGGAUGUGGGUUAGUCCAGGGAACUGACU ......((((((.-......)))))).....((((((((((.((((.----..(----(((....((((......))))...)))).)))).)))).))))))((((.((....)))))) ( -30.40) >consensus CUUAUGCUGUCAGCAUAACAUGGCAGGAAAAAAAUCGGUCCAAAGGA____UGC____UCGAAAGACAAAUAAAAUUGCGGCCGGGACCCUGGGAUGUGGGUAAGUCCAGGGACCUGACU ......((((((........)))))).........(((((..................((....))(((......))).)))))((.((((((.((........)))))))).))..... (-26.60 = -26.32 + -0.28)

| Location | 22,069,059 – 22,069,164 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 82.26 |
| Mean single sequence MFE | -30.67 |
| Consensus MFE | -17.28 |
| Energy contribution | -17.80 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.632588 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
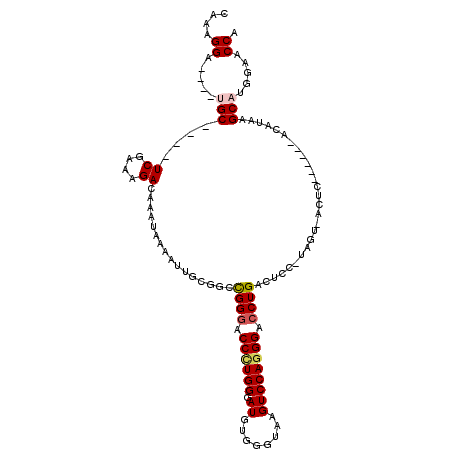
>X_DroMel_CAF1 22069059 105 + 22224390 CAAAGGCUGGCUGC----UCGAAAGACAAAUAAAAUUGCGGCCGGGACCCUGGAAUGUGGGUAAGUCCAGGGACCUGACUCCGUAGUUACUC------ACAUAAGCAUGGAACCA ...(((((((((((----((....))...........))))))))..(((((((.(.......).))))))).)))...(((((.(((....------.....)))))))).... ( -33.60) >DroSec_CAF1 127042 89 + 1 CAAAGGA----UGC----UCGAAAGACAAAUAAAAUUGCGGCCGGGACCCUGGUAUGUGGGUAAGUCCAGGGACCUGAC-----------UC------ACAUAAGCAUG-AACCA ....((.----((.----((....))))........(((.(((((....)))))((((((((..(((....)))...))-----------))------))))..)))..-..)). ( -29.30) >DroSim_CAF1 105814 89 + 1 CAAAGGA----UGC----UCGAAAGACAAAUAAAAUUGCGGCCGGGACCCUGGUAUGUGGGUAAGUCCAGGGACCUGAC-----------UC------ACAUAAGCAUG-AACCA ....((.----((.----((....))))........(((.(((((....)))))((((((((..(((....)))...))-----------))------))))..)))..-..)). ( -29.30) >DroEre_CAF1 101482 105 + 1 CAAAGGA----CGCUCGCUCGAAAGACAAAUAAAAUUGCGGCCGGGACCGUGGGAUGUGGGCUAGUCCAGGGACCUGACUCCAUAGUCACUC------ACAUAAGCAUGGAACCA ....((.----(((....((....))...........))).))((..(((((..(((((((...(((....))).(((((....))))))))------))))...)))))..)). ( -32.36) >DroYak_CAF1 110559 107 + 1 CAAAGGA----CGC----UCAAAAGACAAAUAAAAUUGUGGCUGGGACCUUGGGAUGUGGGUUAGUCCAGGGAACUGACUCCAUAGUCACUCAUAGUCACAUAAGCAUGGCACCA ....((.----.((----.((....((((......)))).(((.((((..((((.((((((((((((....).)))))).)))))....))))..))).)...))).)))).)). ( -28.80) >consensus CAAAGGA____UGC____UCGAAAGACAAAUAAAAUUGCGGCCGGGACCCUGGGAUGUGGGUAAGUCCAGGGACCUGACUCC_UAGU_ACUC______ACAUAAGCAUGGAACCA ....((.....(((....((....))................((((.((((((.((........)))))))).))))...........................))).....)). (-17.28 = -17.80 + 0.52)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:10:24 2006