
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 22,020,463 – 22,020,553 |
| Length | 90 |
| Max. P | 0.614436 |

| Location | 22,020,463 – 22,020,553 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 80.24 |
| Mean single sequence MFE | -28.67 |
| Consensus MFE | -17.76 |
| Energy contribution | -18.32 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.614436 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
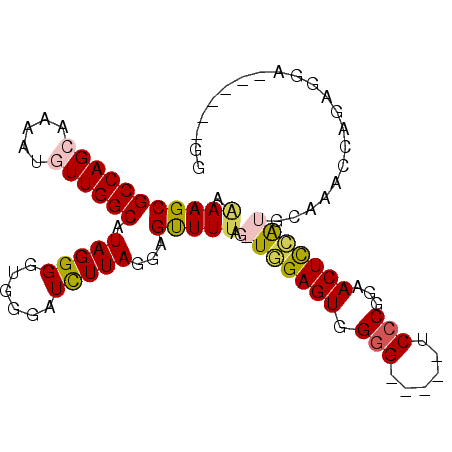
>X_DroMel_CAF1 22020463 90 - 22224390 AGAAGCGCCAGGAAAAUGUUGGCAUAGGGGCGAGAUUUUAGGGGUUUUAGGUGGAGUGGGG-----UCCCGGAACUUCAUGCAAACCAGGGGA------GA .(((((.((...(((((.(((.(.....).))).))))).)).))))).(((...((((((-----(......)))))))....)))......------.. ( -23.40) >DroSec_CAF1 83102 89 - 1 AAAAGCGCCAGCAAAAUGUUGGCAUAGGGGUGGGAUCUUAUGAGUUUUAG-UUGAGUGGGG-----UCGCGGAACUCAAUGCAAACCAGAGGA------GG ....((((((((.....))))))...(((.((.((((((((..(......-.)..))))))-----)).))...)))...))...........------.. ( -23.60) >DroSim_CAF1 61711 95 - 1 AAAAGCGCCAGCAAAAUGUUGGCAUAGGGGUGGGAUCUUAUGAGUUUUAGGUUGAGUGGGGUUGGGUCCCGGAACUCCAUGCAAACCAGAGGA------GG .(((((((((((.....))))))((((((......))))))..))))).((((..((((((((.(....).).)))))))...))))......------.. ( -30.60) >DroEre_CAF1 57235 94 - 1 AGAAGCGCCAGGAAAAUGUUGGCAUAGGGGUGGCAUUUUAGGGGCUUAAA-UGGAGUGGGG-----UCCCGGGACUCCGCGCA-UCCGGGGGAUCCGGGGG ....(((((...(((((((((.(.....).)))))))))...))).....-....((((((-----((....)))))))))).-(((((.....))))).. ( -37.10) >consensus AAAAGCGCCAGCAAAAUGUUGGCAUAGGGGUGGGAUCUUAGGAGUUUUAG_UGGAGUGGGG_____UCCCGGAACUCCAUGCAAACCAGAGGA______GG .(((((((((((.....)))))).(((((......)))))...)))))..(((((((.(((......)))...)))))))..................... (-17.76 = -18.32 + 0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:10:17 2006