
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 22,014,816 – 22,014,952 |
| Length | 136 |
| Max. P | 0.814625 |

| Location | 22,014,816 – 22,014,936 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.17 |
| Mean single sequence MFE | -47.02 |
| Consensus MFE | -20.47 |
| Energy contribution | -22.14 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.45 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.792511 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
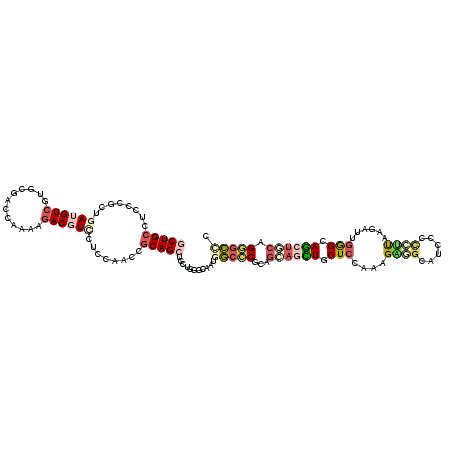
>X_DroMel_CAF1 22014816 120 + 22224390 CCUGCCUCCCGCUGAUGUCGUGCGUCCAAAAGACGUUCUCCAAUCGCAGCUCUUUGGCAAUGGCCCGUAGCAGUUUCUCCAAAGAGGCAUCCCCCUUAAGAUUGAGCAGCUGCAGGGCCC ..((((....((((.((..(.(((((.....))))).)..))....)))).....))))..(((((...((((((.(((....((((......))))......))).)))))).))))). ( -41.80) >DroSec_CAF1 82209 120 + 1 CCUGCCUCCCGCUGAUGUCGUGCGUCCAAAAGACGUUCUCCAAUCGCAGCUCUUUGGCAAUGGCCCGCAGCAGCUUCUCCAAAGAAGCAUCCCCCUUAAGAUUGGGCAGCUGCAGGGCCC ..((((....((((.((..(.(((((.....))))).)..))....)))).....))))..((((((((((.((((((....))))))....(((........)))..))))).))))). ( -49.50) >DroEre_CAF1 56353 120 + 1 GCUGCCUCCCGCUGAUGUCGUGCGACCAGAAGACGUCCUCCAAGCGCAGCUCCUGGGCAAUGGCCCUCAAGAGCUGCUCCAAAGGCGCGUCCUCCCCAAGAUUGGGCAGCUGCAGGGCCC ...((((..(((((..(((....)))..((.((((((((....(.(((((((..((((....))))....))))))).)...))).))))).))((((....)))))))).)..)))).. ( -47.10) >DroYak_CAF1 64985 120 + 1 GCUGCCUCCCGCCGAUGUCGUGCGACCAGAAGACGUCCUCCAACCGCAGCUCCUGGGCAAUGGCCCUCAGCAGCUGCUCCAACGAGGCAUCCUCCUUAAGGUUGGGCAGCUGCAGGGCUC (((((........((((((...(.....)..))))))........)))))((((((((....))))...((((((((.(((((((((......))))...)))))))))))))))))... ( -51.99) >DroMoj_CAF1 34465 120 + 1 GCUGGCCGCCGCUGAGGUCCUGUGACCAGAACACAUGCUCCAGCCGCAGCUGGGAGAUAAUCUGGCGCAGCACCUUCUCCAUUGGCGCUGUCCGGUCCACAUCCGGCAGCUCCAGCGAUC (((((..((((.((((((.(((((.(((((.......(((((((....))).))))....)))))))))).))))....)).))))((((.((((.......)))))))).))))).... ( -55.60) >DroAna_CAF1 33797 120 + 1 GCUGCUUGCCCUCCACGUCCUGCGACCAAAAGACAUCCUCCAGCCGCAGCUCGUCCACAAUCGCCCGCAGGACGUGCUCCAGCGUGGCAUUGUUCUUCAGAUAGGGCAUCACCAGAGCUC .(((..((((((..(((..(((((....................)))))..)))..(((((.((((((.(((.....))).))).)))))))).........))))))....)))..... ( -36.15) >consensus GCUGCCUCCCGCUGAUGUCGUGCGACCAAAAGACGUCCUCCAACCGCAGCUCCUGGGCAAUGGCCCGCAGCAGCUGCUCCAAAGAGGCAUCCCCCUUAAGAUUGGGCAGCUGCAGGGCCC (((((........((((((............))))))........)))))...........(((((...((((((.(((....((((......))))......))).)))))).))))). (-20.47 = -22.14 + 1.67)

| Location | 22,014,856 – 22,014,952 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 67.73 |
| Mean single sequence MFE | -37.43 |
| Consensus MFE | -15.03 |
| Energy contribution | -16.95 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.67 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.40 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.814625 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 22014856 96 + 22224390 CAAUCGCAGCUCUUUGGCAAUGGCCCGUAGCAGUUUCUCCAAAGAGGCAUCCCCCUUAAGAUUGAGCAGCUGCAGGGCCCUCCCCUGGGACAUAGG ........(((....)))...(((((...((((((.(((....((((......))))......))).)))))).))))).(((....)))...... ( -29.30) >DroVir_CAF1 33363 94 + 1 CAGCCGCAACUGCAGGGCAAUCUGGCGUACCACCUGCUCCAGUGGUGCAUUUCUUGACAGCUCCCGCAGGUGCAGAGCUCGGCUGUUGU--GUGAC ((((((((.((((.((((..((.((.(((((((........)))))))....)).))..)).)).)))).))).(....))))))....--..... ( -34.10) >DroGri_CAF1 37427 94 + 1 CAGCCGCAGUUGCAUUACAAUCCGACGCAGCACUUGUGCCAGCGGUGUAUUGUCCGGCAACUCCGGCAGCUGCAGCGCUCUACUCAUCC--GUGGC ..((((.((((((...(((((.((.(((.(((....)))..))).)).)))))...)))))).)))).((....))...((((......--)))). ( -32.80) >DroSec_CAF1 82249 96 + 1 CAAUCGCAGCUCUUUGGCAAUGGCCCGCAGCAGCUUCUCCAAAGAAGCAUCCCCCUUAAGAUUGGGCAGCUGCAGGGCCCUGCUCUGGGACAUAGG ........(..((..((((..((((((((((.((((((....))))))....(((........)))..))))).))))).))))..))..)..... ( -39.80) >DroEre_CAF1 56393 96 + 1 CAAGCGCAGCUCCUGGGCAAUGGCCCUCAAGAGCUGCUCCAAAGGCGCGUCCUCCCCAAGAUUGGGCAGCUGCAGGGCCCUGCUCUGGCUCCUAAG .....(((((((..((((....))))....))))))).....(((.((......((((....))))(((..((((....)))).))))).)))... ( -40.10) >DroYak_CAF1 65025 96 + 1 CAACCGCAGCUCCUGGGCAAUGGCCCUCAGCAGCUGCUCCAACGAGGCAUCCUCCUUAAGGUUGGGCAGCUGCAGGGCUCUGCUCUGGCACUUUGG ...(((.((..((.(((((..((((((..((((((((.(((((((((......))))...))))))))))))))))))).))))).))..)).))) ( -48.50) >consensus CAACCGCAGCUCCUGGGCAAUGGCCCGCAGCAGCUGCUCCAAAGAGGCAUCCCCCUUAAGAUUGGGCAGCUGCAGGGCCCUGCUCUGGCACAUAGG ...........((((((....(((((...((((((((((.....(((.......)))......)))))))))).)))))....))))))....... (-15.03 = -16.95 + 1.92)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:10:16 2006