
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 21,996,962 – 21,997,078 |
| Length | 116 |
| Max. P | 0.790193 |

| Location | 21,996,962 – 21,997,078 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 81.78 |
| Mean single sequence MFE | -35.60 |
| Consensus MFE | -24.68 |
| Energy contribution | -24.59 |
| Covariance contribution | -0.09 |
| Combinations/Pair | 1.40 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.790193 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
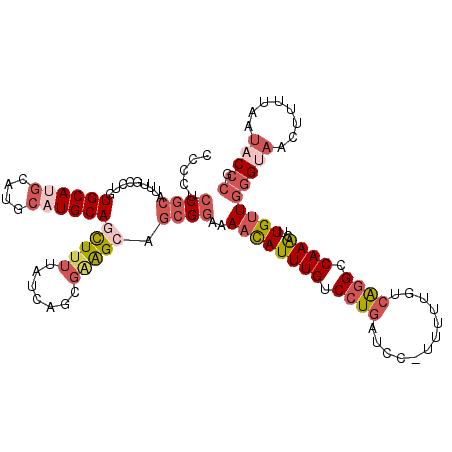
>X_DroMel_CAF1 21996962 116 + 22224390 CGGGUAUUAAAAGUUACCCAACAAGUUUGGCCUGACAAAAA-GGAUCAGGACAAAUGUUUUCCGCUGCUUCGCUGAUAAAAGCUGCAUGCAUGCAUGCACAGGCAAAUGCGGUGGGG .(((((........))))).....(((((.((((((.....-.).))))).)))))....((((((((...(((......)))((((((....)))))).........)))))))). ( -40.70) >DroGri_CAF1 20171 102 + 1 -----------AGUUG-CCAGCAAACUUGGCACUCCAAAAA-CGACCAAGACAAAUAUUUU-CGUUGCCUGUGUGAUAAAAACUGCAUGCGAGUGUGCAUUG-UAUGUGUGUAUGUG -----------(((((-((((.....)))))..((((....-((((.((((......))))-.))))....)).))....))))(((((((.(..(((...)-))..).))))))). ( -19.80) >DroSec_CAF1 57542 116 + 1 CGGGUAUUAAAAGUUACCCAACAAGUUUGGCCUGACAAAAA-GGAUCAGGACAAAUGUUUUCCGCUGCUCCGUUGAUAAAAGCUGCAUGCAUGCAUGCACAGGCAAAUGCGGAGGGG .(((((........))))).....(((((.((((((.....-.).))))).))))).((((((((((((..(((......)))((((((....))))))..))))...)))))))). ( -38.40) >DroSim_CAF1 57567 116 + 1 CGGGUAUUAAAAGUUACCCAACAAGUUUGGCCUGACAAAAA-GGAUCAGGACAAAUGUUUUCCGCUGCUCCGCUGAUAAAAGCUGCAUGCAUGCAUGCACAGGCAAAUGCGGAGGGG .(((((........))))).....(((((.((((((.....-.).))))).))))).((((((((((((..(((......)))((((((....))))))..))))...)))))))). ( -40.80) >DroEre_CAF1 37863 114 + 1 CGGGUAUUAAAAGUUGCCCAACAAGUUUGGCCUGGCAAAA--GGAUCAGGACAAAUGUUUUCCGCUGCUUCGCUGAUAAAAAUUGCAUGCGUGCGUGCACU-GAAAAUGCGGUGGGG .(((((........))))).....(((((.(((((.....--...))))).)))))....((((((((...............((((((....)))))).(-....).)))))))). ( -34.10) >DroYak_CAF1 46313 117 + 1 CGGGUAUUAAAAGUUACCCAACAAGUUUGGCCUGGCAAAAAGGGAUCAGGACAAAUGUUUUCCGCUGCUUCUCUGAUUAGAAUUGCAUGCAUGCAUGCAGAGGGAAAUGCGGAGGAG .(((((........))))).....(((((.((((((.......).))))).))))).((((((((...((((((........(((((((....)))))))))))))..)))))))). ( -39.80) >consensus CGGGUAUUAAAAGUUACCCAACAAGUUUGGCCUGACAAAAA_GGAUCAGGACAAAUGUUUUCCGCUGCUUCGCUGAUAAAAACUGCAUGCAUGCAUGCACAGGCAAAUGCGGAGGGG .(((((........))))).....(((((.((((((.......).))))).)))))...........((((((((........((((((....))))))..........)))))))) (-24.68 = -24.59 + -0.09)



| Location | 21,996,962 – 21,997,078 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 81.78 |
| Mean single sequence MFE | -31.45 |
| Consensus MFE | -19.90 |
| Energy contribution | -21.97 |
| Covariance contribution | 2.07 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.696825 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21996962 116 - 22224390 CCCCACCGCAUUUGCCUGUGCAUGCAUGCAUGCAGCUUUUAUCAGCGAAGCAGCGGAAAACAUUUGUCCUGAUCC-UUUUUGUCAGGCCAAACUUGUUGGGUAACUUUUAAUACCCG .....((((.........((((((....))))))(((((.......))))).))))..((((((((.((((((..-.....)))))).))))..))))(((((........))))). ( -37.20) >DroGri_CAF1 20171 102 - 1 CACAUACACACAUA-CAAUGCACACUCGCAUGCAGUUUUUAUCACACAGGCAACG-AAAAUAUUUGUCUUGGUCG-UUUUUGGAGUGCCAAGUUUGCUGG-CAACU----------- ..............-...(((.((...(((.((.((.......))...((((.((-((((((((......))).)-))))))...))))..)).))))))-))...----------- ( -16.70) >DroSec_CAF1 57542 116 - 1 CCCCUCCGCAUUUGCCUGUGCAUGCAUGCAUGCAGCUUUUAUCAACGGAGCAGCGGAAAACAUUUGUCCUGAUCC-UUUUUGUCAGGCCAAACUUGUUGGGUAACUUUUAAUACCCG ....(((((.........((((((....))))))(((((.......))))).))))).((((((((.((((((..-.....)))))).))))..))))(((((........))))). ( -38.00) >DroSim_CAF1 57567 116 - 1 CCCCUCCGCAUUUGCCUGUGCAUGCAUGCAUGCAGCUUUUAUCAGCGGAGCAGCGGAAAACAUUUGUCCUGAUCC-UUUUUGUCAGGCCAAACUUGUUGGGUAACUUUUAAUACCCG ....(((((...(((((.((((((....))))))(((......))))).)))))))).((((((((.((((((..-.....)))))).))))..))))(((((........))))). ( -38.70) >DroEre_CAF1 37863 114 - 1 CCCCACCGCAUUUUC-AGUGCACGCACGCAUGCAAUUUUUAUCAGCGAAGCAGCGGAAAACAUUUGUCCUGAUCC--UUUUGCCAGGCCAAACUUGUUGGGCAACUUUUAAUACCCG .(((.((((..((((-.((((......))))((...........))))))..))))..((((((((.((((....--......)))).))))..)))))))................ ( -25.50) >DroYak_CAF1 46313 117 - 1 CUCCUCCGCAUUUCCCUCUGCAUGCAUGCAUGCAAUUCUAAUCAGAGAAGCAGCGGAAAACAUUUGUCCUGAUCCCUUUUUGCCAGGCCAAACUUGUUGGGUAACUUUUAAUACCCG ....(((((.........((((((....)))))).((((......))))...))))).((((((((.((((............)))).))))..))))(((((........))))). ( -32.60) >consensus CCCCUCCGCAUUUGCCUGUGCAUGCAUGCAUGCAGCUUUUAUCAGCGAAGCAGCGGAAAACAUUUGUCCUGAUCC_UUUUUGUCAGGCCAAACUUGUUGGGUAACUUUUAAUACCCG .....((((.........((((((....))))))(((((.......))))).))))..((((((((.((((............)))).))))..))))(((((........))))). (-19.90 = -21.97 + 2.07)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:10:12 2006