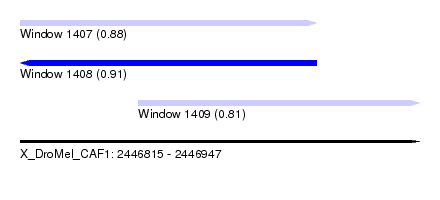
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,446,815 – 2,446,947 |
| Length | 132 |
| Max. P | 0.905250 |

| Location | 2,446,815 – 2,446,913 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.94 |
| Mean single sequence MFE | -45.77 |
| Consensus MFE | -34.71 |
| Energy contribution | -35.93 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.882941 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2446815 98 + 22224390 UUUACUCGGCGGACUGCUCUGUGCAAUCCAGUUGUCCAU---UUGCCCUAAUCGAAGGGGA-UGGGCCAACCGUUUGCAUGGCC-CCUUUUGUAACUGCCGGC----------------- .....((((((((.(((.....))).)))(((((.....---...((((......))))..-.((((((..........)))))-)......)))))))))).----------------- ( -33.40) >DroSec_CAF1 7616 120 + 1 UUUACUCGGCGGACUGCUCUGUGCGGUCCAGUUGUCCAGUUCUUGCCCUAAUCGCAGGGAUGGGGGCCCACCGUUUGCAUGGCCCCCUUUUGCAACUGGCGGCCUGCACUCGAGGAAACC ....(((((((((((((.....))))))).((((.((((((....((((......))))..(((((((............)))))))......))))))))))..))...))))...... ( -50.20) >DroSim_CAF1 9838 120 + 1 UUUACUCGGCGGACUGCUCUGUGCGGUCCAGUUGUCCAGUUCUUGCCCUAAUCGAAGGGCUGGGGGCCCACCGUUUGCAUGGCCCCCUUUUGCAACUGGCGGCCUGCACUCGAGGAAACC ....(((((((((((((.....))))))).((((.((((((...(((((......))))).(((((((............)))))))......))))))))))..))...))))...... ( -53.70) >consensus UUUACUCGGCGGACUGCUCUGUGCGGUCCAGUUGUCCAGUUCUUGCCCUAAUCGAAGGGAUGGGGGCCCACCGUUUGCAUGGCCCCCUUUUGCAACUGGCGGCCUGCACUCGAGGAAACC .......((((((((((.....)))))))((((((..........((((......))))..(((((((............)))))))....))))))....)))................ (-34.71 = -35.93 + 1.23)

| Location | 2,446,815 – 2,446,913 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.94 |
| Mean single sequence MFE | -45.70 |
| Consensus MFE | -30.42 |
| Energy contribution | -33.20 |
| Covariance contribution | 2.78 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.905250 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2446815 98 - 22224390 -----------------GCCGGCAGUUACAAAAGG-GGCCAUGCAAACGGUUGGCCCA-UCCCCUUCGAUUAGGGCAA---AUGGACAACUGGAUUGCACAGAGCAGUCCGCCGAGUAAA -----------------((((((((((.......(-(((((.((.....)))))))).-..((((......))))...---......))))(((((((.....))))))))))).))... ( -38.10) >DroSec_CAF1 7616 120 - 1 GGUUUCCUCGAGUGCAGGCCGCCAGUUGCAAAAGGGGGCCAUGCAAACGGUGGGCCCCCAUCCCUGCGAUUAGGGCAAGAACUGGACAACUGGACCGCACAGAGCAGUCCGCCGAGUAAA ((.((.(((..((((.((...((((((((....(((((((.(((.....))))))))))..(((((....))))).........).))))))).)))))).))).)).)).......... ( -47.20) >DroSim_CAF1 9838 120 - 1 GGUUUCCUCGAGUGCAGGCCGCCAGUUGCAAAAGGGGGCCAUGCAAACGGUGGGCCCCCAGCCCUUCGAUUAGGGCAAGAACUGGACAACUGGACCGCACAGAGCAGUCCGCCGAGUAAA ((.((.(((..((((.((...((((((((....(((((((.(((.....)))))))))).(((((......)))))........).))))))).)))))).))).)).)).......... ( -51.80) >consensus GGUUUCCUCGAGUGCAGGCCGCCAGUUGCAAAAGGGGGCCAUGCAAACGGUGGGCCCCCACCCCUUCGAUUAGGGCAAGAACUGGACAACUGGACCGCACAGAGCAGUCCGCCGAGUAAA ................(((..((((((......(((((((.(((.....))))))))))..((((......))))....))))))......((((.((.....)).)))))))....... (-30.42 = -33.20 + 2.78)

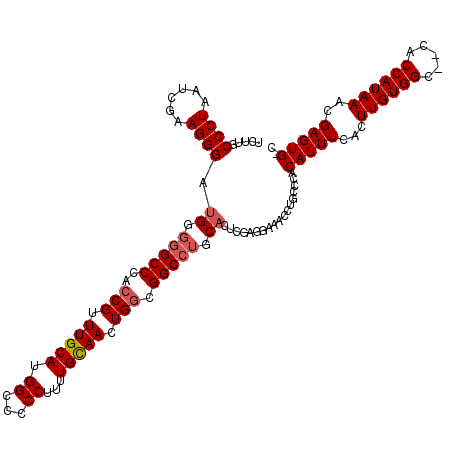
| Location | 2,446,854 – 2,446,947 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.99 |
| Mean single sequence MFE | -41.27 |
| Consensus MFE | -24.30 |
| Energy contribution | -25.50 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.813495 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2446854 93 + 22224390 --UUGCCCUAAUCGAAGGGGA-UGGGCCAACCGUUUGCAUGGCC-CCUUUUGUAACUGCCGGC---------------------CACAUUUUACUUGUGGC--CACCAUAAGCGAGUGGC --..((((...((....))..-.))))...((((((((..(((.-............)))(((---------------------((((.......))))))--).......)))))))). ( -34.22) >DroSec_CAF1 7656 119 + 1 UCUUGCCCUAAUCGCAGGGAUGGGGGCCCACCGUUUGCAUGGCCCCCUUUUGCAACUGGCGGCCUGCACUCGAGGAAACCUGCCCACAUUUCACUUGUGGCCCCACCAUAAACGAGUG-C ....((((((...(((((...(((((((............))))))).)))))...))).)))..(((((((.(....)....(((((.......)))))............))))))-) ( -43.30) >DroSim_CAF1 9878 117 + 1 UCUUGCCCUAAUCGAAGGGCUGGGGGCCCACCGUUUGCAUGGCCCCCUUUUGCAACUGGCGGCCUGCACUCGAGGAAACCUGCCCACAUUUCACUUGUGGC--CACCAUAAACGAGUG-C .((.(((((......))))).))(((((..(((.(((((.((...))...))))).))).)))))(((((((.(....).((.(((((.......))))).--)).......))))))-) ( -46.30) >consensus UCUUGCCCUAAUCGAAGGGAUGGGGGCCCACCGUUUGCAUGGCCCCCUUUUGCAACUGGCGGCCUGCACUCGAGGAAACCUGCCCACAUUUCACUUGUGGC__CACCAUAAACGAGUG_C .....((((......)))).((.(((((..(((.(((((.((...))...))))).))).))))).))..................(((((...((((((.....))))))..))))).. (-24.30 = -25.50 + 1.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:55:50 2006