
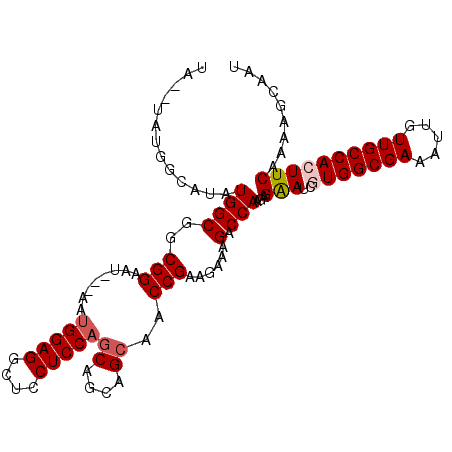
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 21,993,756 – 21,993,858 |
| Length | 102 |
| Max. P | 0.559660 |

| Location | 21,993,756 – 21,993,858 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 87.88 |
| Mean single sequence MFE | -30.26 |
| Consensus MFE | -21.04 |
| Energy contribution | -21.68 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.559660 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21993756 102 + 22224390 UA--UAUGGCAUAUGGCGGCGGAAU---AAUGGAGGCUCCUCCAGCAACAGCAACCGAAGAAAGACCAAGAGAAUCGUGGCGAAAUUGUUGCCACUUCAAAAGCAAU ..--....((...((((..(((...---..(((((....)))))((....))..)))......).)))...(((..(((((((.....))))))))))....))... ( -27.90) >DroSec_CAF1 54308 102 + 1 UG--UAUGUCAUAUGGCGGCGGAAU---AAUGGAGGCUGCUCCAGCAGCAGCAACCGAAGAAAGACCAAGAGAAUCGUGGCGAAAUUGUUGCCACUUCAAAAGCAAU ..--..((((....))))((.(((.---..(((..((((((.....))))))..)))...................(((((((.....))))))))))....))... ( -27.90) >DroSim_CAF1 54341 102 + 1 UG--UAUGUCAUAUGGCGGCGGAAU---AAUGGAGGCUGCUCCAGCAGCAGCAACCGAAGAAAGACCAAGAGAAUCGUGGCGAAAUUGUUGCCACUUCAAAAGCAAU ..--..((((....))))((.(((.---..(((..((((((.....))))))..)))...................(((((((.....))))))))))....))... ( -27.90) >DroEre_CAF1 34879 98 + 1 UAUGUAUGGCAUGUGGCGGCGGAGG---GGAGGAGGCUCCUCCAGCAGCAG---CCGAAGAAAGGCCAAGAGAAACAUGGCGAAAUUGUUGCCACUUCAAAAGC--- ......((((...((((.(((((((---((......))))))).))....)---))).......))))...(((...((((((.....)))))).)))......--- ( -33.30) >DroYak_CAF1 42790 105 + 1 UA--UAUGGCAUGUGGCGGCGGAUGAAGAAUGGAGGCUCCUCCAGCAGCAGCUACCGAAGAAAGGCCAAGAGGAAGAUGGCGAAAUUGUUGCCACUUCAAAAGCAAU ..--..((((...(..(((..(.((.....(((((....)))))....)).)..)))..)....))))....((((.((((((.....))))))))))......... ( -34.30) >consensus UA__UAUGGCAUAUGGCGGCGGAAU___AAUGGAGGCUCCUCCAGCAGCAGCAACCGAAGAAAGACCAAGAGAAUCGUGGCGAAAUUGUUGCCACUUCAAAAGCAAU .............((((..(((........(((((....)))))((....))..)))......).)))...(((..(((((((.....))))))))))......... (-21.04 = -21.68 + 0.64)


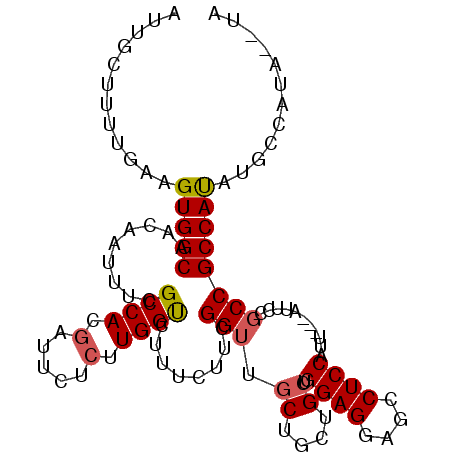
| Location | 21,993,756 – 21,993,858 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 87.88 |
| Mean single sequence MFE | -27.00 |
| Consensus MFE | -20.72 |
| Energy contribution | -20.84 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.547124 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21993756 102 - 22224390 AUUGCUUUUGAAGUGGCAACAAUUUCGCCACGAUUCUCUUGGUCUUUCUUCGGUUGCUGUUGCUGGAGGAGCCUCCAUU---AUUCCGCCGCCAUAUGCCAUA--UA ............((((((........((((.(.....).))))........(((.((.(.((.(((((....))))).)---)..).)).)))...)))))).--.. ( -26.20) >DroSec_CAF1 54308 102 - 1 AUUGCUUUUGAAGUGGCAACAAUUUCGCCACGAUUCUCUUGGUCUUUCUUCGGUUGCUGCUGCUGGAGCAGCCUCCAUU---AUUCCGCCGCCAUAUGACAUA--CA ............(((((.........((((.(.....).))))........((..((((((.....))))))..))...---.....)))))...........--.. ( -25.50) >DroSim_CAF1 54341 102 - 1 AUUGCUUUUGAAGUGGCAACAAUUUCGCCACGAUUCUCUUGGUCUUUCUUCGGUUGCUGCUGCUGGAGCAGCCUCCAUU---AUUCCGCCGCCAUAUGACAUA--CA ............(((((.........((((.(.....).))))........((..((((((.....))))))..))...---.....)))))...........--.. ( -25.50) >DroEre_CAF1 34879 98 - 1 ---GCUUUUGAAGUGGCAACAAUUUCGCCAUGUUUCUCUUGGCCUUUCUUCGG---CUGCUGCUGGAGGAGCCUCCUCC---CCUCCGCCGCCACAUGCCAUACAUA ---.........((((((........((((.(.....).))))........((---(.((.(..(((((.....)))))---...).)).)))...))))))..... ( -30.10) >DroYak_CAF1 42790 105 - 1 AUUGCUUUUGAAGUGGCAACAAUUUCGCCAUCUUCCUCUUGGCCUUUCUUCGGUAGCUGCUGCUGGAGGAGCCUCCAUUCUUCAUCCGCCGCCACAUGCCAUA--UA ............(((((.........((((.........))))........((((((....)))((((....))))...........))))))))........--.. ( -27.70) >consensus AUUGCUUUUGAAGUGGCAACAAUUUCGCCACGAUUCUCUUGGUCUUUCUUCGGUUGCUGCUGCUGGAGGAGCCUCCAUU___AUUCCGCCGCCAUAUGCCAUA__UA ............(((((.........((((.(.....).))))........(((.((....)).((((....))))...........))))))))............ (-20.72 = -20.84 + 0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:10:10 2006