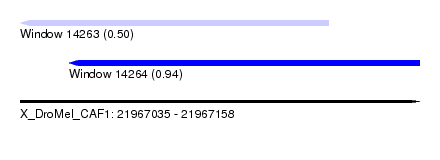
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 21,967,035 – 21,967,158 |
| Length | 123 |
| Max. P | 0.944830 |

| Location | 21,967,035 – 21,967,130 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.24 |
| Mean single sequence MFE | -35.52 |
| Consensus MFE | -27.63 |
| Energy contribution | -28.11 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.78 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21967035 95 - 22224390 AUGCAACCAGUACAUCCUGUUGUCGUUGUGGUCAGAUUUCCGAUUGCUGCAAUCGGAUGAACAGUCGGCGACAGGGUGCCUUAA-------------------------AUGCGGGUAGG .....(((.((((((((((((((((((((..(((....((((((((...))))))))))))))).)))))))))))))......-------------------------.))).)))... ( -36.41) >DroSec_CAF1 32139 95 - 1 AUGCAACCACUACAUCCUGUUGUCGUUGUCGUCAGAUUUCCGAUUGCUGCAAUCGGAUGAACAGUCGUCGUCAGGGGGCCUUAA-------------------------AUGGGGGUAGG .........((((.(((((.((.((((((..(((....((((((((...))))))))))))))).)).)).)))))..((((..-------------------------..)))))))). ( -28.40) >DroSim_CAF1 18832 95 - 1 AUGCAACCACUACAUCCUGUUGUCGUUGUCGUCAGAUUUCCGAUUGCUGCAAUCGGAUGAACAGUCGUCGUCAGGGGGCCUUAA-------------------------AUGGGGGUAGG .........((((.(((((.((.((((((..(((....((((((((...))))))))))))))).)).)).)))))..((((..-------------------------..)))))))). ( -28.40) >DroEre_CAF1 14741 120 - 1 AUGCAACCAGUACAUCCUGUUGUCGUUGUCAUCAGAUUUCCGAUUGCUGCAAUCGGAUGAACAGUCGGCGACAGAGGACCUCAGGCGAGAGGGGCAGGCGACAGGGGGGAAGGGGGCAGG .(((..((..(...((((((((((((((((..(.....((((((((...))))))))......)..))))))...(..((((......))))..).))))))))))..)..))..))).. ( -47.30) >DroYak_CAF1 21773 95 - 1 AUGCAACCAGUACAUCCUGUUGUCGUUGUCGUCAGACUUCCGAUUGCUGCAAUCGGAUGAGCAGUCGGCGGCAGGGGACCUCAA-------------------------AAGGGGGCAGG .(((..........(((((((((((((((..(((....((((((((...))))))))))))))).)))))))))))..((((..-------------------------..))))))).. ( -37.10) >consensus AUGCAACCAGUACAUCCUGUUGUCGUUGUCGUCAGAUUUCCGAUUGCUGCAAUCGGAUGAACAGUCGGCGACAGGGGGCCUUAA_________________________AUGGGGGUAGG .(((..........(((((((((((((((..(((....((((((((...))))))))))))))).)))))))))))..((((.............................))))))).. (-27.63 = -28.11 + 0.48)

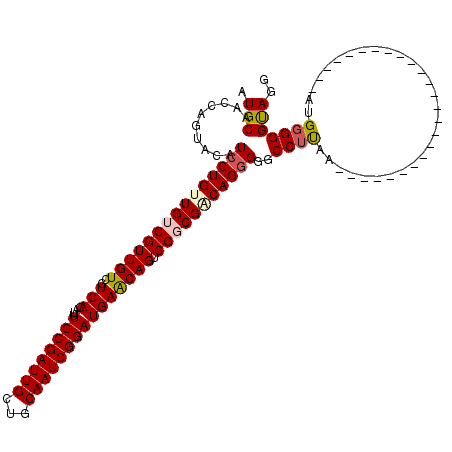

| Location | 21,967,050 – 21,967,158 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.49 |
| Mean single sequence MFE | -36.92 |
| Consensus MFE | -28.26 |
| Energy contribution | -29.18 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.944830 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21967050 108 - 22224390 UCC------------UUGUUCAUUAAAUUAGGUGCAUUAAAUGCAACCAGUACAUCCUGUUGUCGUUGUGGUCAGAUUUCCGAUUGCUGCAAUCGGAUGAACAGUCGGCGACAGGGUGCC ...------------...............(((((((...)))).)))....(((((((((((((((((..(((....((((((((...))))))))))))))).))))))))))))).. ( -39.40) >DroSec_CAF1 32154 120 - 1 UCCCUUGGCAACCCUUUGUUAAUUAAAUUAAGUGCAUUAAAUGCAACCACUACAUCCUGUUGUCGUUGUCGUCAGAUUUCCGAUUGCUGCAAUCGGAUGAACAGUCGUCGUCAGGGGGCC (((((((((.((...(((((............(((((...))))).......(((((.(((((.((.((((.........)))).)).))))).))))))))))..)).))))))))).. ( -37.30) >DroSim_CAF1 18847 120 - 1 UCCCUUGGAAACCCUUUGUUCAUUAAAUUAAGUGCAUUAAAUGCAACCACUACAUCCUGUUGUCGUUGUCGUCAGAUUUCCGAUUGCUGCAAUCGGAUGAACAGUCGUCGUCAGGGGGCC ((((((((..((...((((((...........(((((...)))))........((((.(((((.((.((((.........)))).)).))))).))))))))))..))..)))))))).. ( -35.10) >DroEre_CAF1 14781 119 - 1 UCCCUU-GCUACCCUUUGUUCAUUAAAUUAGCUGCAUUAAAUGCAACCAGUACAUCCUGUUGUCGUUGUCAUCAGAUUUCCGAUUGCUGCAAUCGGAUGAACAGUCGGCGACAGAGGACC ..(((.-((((...((((.....)))).))))(((((...)))))...........(((((((((((((..(((....((((((((...))))))))))))))).))))))))))))... ( -32.40) >DroYak_CAF1 21788 120 - 1 UCCCUUGGCUACCAUUUGUUCAUUAAAUUAGCUGCAUUAAAUGCAACCAGUACAUCCUGUUGUCGUUGUCGUCAGACUUCCGAUUGCUGCAAUCGGAUGAGCAGUCGGCGGCAGGGGACC (((((((.(..((..(((((((.......(((((.....((((((((.((......)))))).)))).....))).))((((((((...)))))))))))))))..))..)))))))).. ( -40.40) >consensus UCCCUUGGCAACCCUUUGUUCAUUAAAUUAGGUGCAUUAAAUGCAACCAGUACAUCCUGUUGUCGUUGUCGUCAGAUUUCCGAUUGCUGCAAUCGGAUGAACAGUCGGCGACAGGGGGCC .................((((...........(((((...)))))..........((((((((((((((..(((....((((((((...))))))))))))))).)))))))))))))). (-28.26 = -29.18 + 0.92)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:10:05 2006