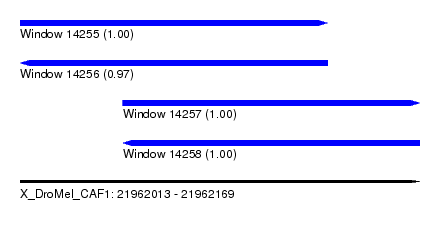
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 21,962,013 – 21,962,169 |
| Length | 156 |
| Max. P | 0.999941 |

| Location | 21,962,013 – 21,962,133 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.50 |
| Mean single sequence MFE | -25.00 |
| Consensus MFE | -23.26 |
| Energy contribution | -23.70 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.32 |
| Structure conservation index | 0.93 |
| SVM decision value | 4.71 |
| SVM RNA-class probability | 0.999941 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21962013 120 + 22224390 UAAUCGACAAAACACACAGAAACACCCUAGGCCGGUUCACACAUACCACCAGACAUAUUGCGUAUACGCUGACCAUAUUGCGUAUACUCAACCAACACAAUAGCUCCGCCAGCCAUCCAA .............................(((.(((........)))..........(((.((((((((..........)))))))).)))................))).......... ( -20.50) >DroSec_CAF1 24395 120 + 1 UAAUCGACAAAACACACAGAAACACCCUAGGCCGGCUCACACAUACCACCAGACAUAUUGCGUAUACGCUGACCAUAUUGCGUAUACGCAACCAACACAAUAGCUCCGCCAGCCAUCCAA .............................(((.(((..........(....).....((((((((((((..........))))))))))))................))).)))...... ( -27.60) >DroSim_CAF1 13611 120 + 1 UAAUCGACAAAACACACAGAAACACCCUAGGCCGGCUCACACAUACCACCAGACAUAUUGCGUAUACGCUGACCAUAUUGCGUAUACGCAACCAACACAAUAGCUCCGCCAGCCAUCCAA .............................(((.(((..........(....).....((((((((((((..........))))))))))))................))).)))...... ( -27.60) >DroEre_CAF1 9653 120 + 1 UAAUCGACAAAACACACAGAAACACCCUAGGCCGGCUCACACACACCGCCACACAUAUUGCGUAUACGCUGGCCAUAUUGCGUAUACGCCACCAACACAAUAGCUCCGCCAGCCAGCCAA .............................(((.((((......................((((((((((..........)))))))))).............((...)).)))).))).. ( -27.00) >DroYak_CAF1 16723 120 + 1 UAAUCGACAAAACACACAGAAACACCCUAGGCCGCCUCACACAUACCGCCACACAUAUUGCGUAUACGCUGGCCAUAUUGCGUAUACGCAACCAACACAAUAGCUCCGCCAGCCAUCCAA .............................(((.((............))........((((((((((((..........))))))))))))................))).......... ( -22.30) >consensus UAAUCGACAAAACACACAGAAACACCCUAGGCCGGCUCACACAUACCACCAGACAUAUUGCGUAUACGCUGACCAUAUUGCGUAUACGCAACCAACACAAUAGCUCCGCCAGCCAUCCAA .............................(((.(((.....................((((((((((((..........))))))))))))....(......)....))).)))...... (-23.26 = -23.70 + 0.44)

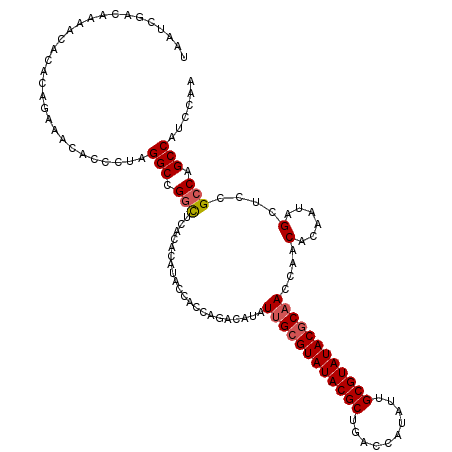

| Location | 21,962,013 – 21,962,133 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.50 |
| Mean single sequence MFE | -38.96 |
| Consensus MFE | -33.52 |
| Energy contribution | -33.72 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.965890 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21962013 120 - 22224390 UUGGAUGGCUGGCGGAGCUAUUGUGUUGGUUGAGUAUACGCAAUAUGGUCAGCGUAUACGCAAUAUGUCUGGUGGUAUGUGUGAACCGGCCUAGGGUGUUUCUGUGUGUUUUGUCGAUUA ((((..(((..((((((.((((.((..(((((.((.((((((.(((..(((((((((.....))))).))))..))))))))).))))))))).)))).))))))..)))...))))... ( -33.80) >DroSec_CAF1 24395 120 - 1 UUGGAUGGCUGGCGGAGCUAUUGUGUUGGUUGCGUAUACGCAAUAUGGUCAGCGUAUACGCAAUAUGUCUGGUGGUAUGUGUGAGCCGGCCUAGGGUGUUUCUGUGUGUUUUGUCGAUUA ..((.(((((.(((.(((((((......(((((((((((((..........)))))))))))))......)))))).).))).))))).))(((((..(......)..)))))....... ( -39.00) >DroSim_CAF1 13611 120 - 1 UUGGAUGGCUGGCGGAGCUAUUGUGUUGGUUGCGUAUACGCAAUAUGGUCAGCGUAUACGCAAUAUGUCUGGUGGUAUGUGUGAGCCGGCCUAGGGUGUUUCUGUGUGUUUUGUCGAUUA ..((.(((((.(((.(((((((......(((((((((((((..........)))))))))))))......)))))).).))).))))).))(((((..(......)..)))))....... ( -39.00) >DroEre_CAF1 9653 120 - 1 UUGGCUGGCUGGCGGAGCUAUUGUGUUGGUGGCGUAUACGCAAUAUGGCCAGCGUAUACGCAAUAUGUGUGGCGGUGUGUGUGAGCCGGCCUAGGGUGUUUCUGUGUGUUUUGUCGAUUA (((((.(((((((...((((((.....))))))...((((((.(((.((((((((((.....)))))).)))).))))))))).)))))))..(((....))).........)))))... ( -42.80) >DroYak_CAF1 16723 120 - 1 UUGGAUGGCUGGCGGAGCUAUUGUGUUGGUUGCGUAUACGCAAUAUGGCCAGCGUAUACGCAAUAUGUGUGGCGGUAUGUGUGAGGCGGCCUAGGGUGUUUCUGUGUGUUUUGUCGAUUA ((((..(((..((((((.((((.((..((((((.(.((((((.(((.((((((((((.....)))))).)))).)))))))))).)))))))).)))).))))))..)))...))))... ( -40.20) >consensus UUGGAUGGCUGGCGGAGCUAUUGUGUUGGUUGCGUAUACGCAAUAUGGUCAGCGUAUACGCAAUAUGUCUGGUGGUAUGUGUGAGCCGGCCUAGGGUGUUUCUGUGUGUUUUGUCGAUUA ((((..(((..((((((.((((.((...(((((((((((((..........)))))))))))))......((((((........))).))))).)))).))))))..)))...))))... (-33.52 = -33.72 + 0.20)



| Location | 21,962,053 – 21,962,169 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.27 |
| Mean single sequence MFE | -22.50 |
| Consensus MFE | -20.30 |
| Energy contribution | -20.70 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.90 |
| SVM decision value | 3.25 |
| SVM RNA-class probability | 0.998849 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21962053 116 + 22224390 ACAUACCACCAGACAUAUUGCGUAUACGCUGACCAUAUUGCGUAUACUCAACCAACACAAUAGCUCCGCCAGCCAUCCAAUGCCAAACCGCUUAAAGCCA----AAAUUGCCAGCGACAA ...........(.(((.(((.((((((((..........)))))))).)))...........(((.....)))......))).)....((((....((..----.....)).)))).... ( -17.90) >DroSec_CAF1 24435 116 + 1 ACAUACCACCAGACAUAUUGCGUAUACGCUGACCAUAUUGCGUAUACGCAACCAACACAAUAGCUCCGCCAGCCAUCCAAUGCCAAACCGCUUAAAGCCA----AAAUUGCCAGCGACAA ...........(.(((.((((((((((((..........))))))))))))...........(((.....)))......))).)....((((....((..----.....)).)))).... ( -23.70) >DroSim_CAF1 13651 116 + 1 ACAUACCACCAGACAUAUUGCGUAUACGCUGACCAUAUUGCGUAUACGCAACCAACACAAUAGCUCCGCCAGCCAUCCAAUGCCAAACCGCUUAAAGCCA----AAAUUGCCAGCGACAA ...........(.(((.((((((((((((..........))))))))))))...........(((.....)))......))).)....((((....((..----.....)).)))).... ( -23.70) >DroEre_CAF1 9693 119 + 1 ACACACCGCCACACAUAUUGCGUAUACGCUGGCCAUAUUGCGUAUACGCCACCAACACAAUAGCUCCGCCAGCCAGCCAAUGCCAAACCGCUUAAAGCCAAC-AAAAUUGCCAGCGACAA ......(((..........((((((((((..........))))))))))........((((.(((.....)))..((....((......)).....))....-...))))...))).... ( -22.60) >DroYak_CAF1 16763 120 + 1 ACAUACCGCCACACAUAUUGCGUAUACGCUGGCCAUAUUGCGUAUACGCAACCAACACAAUAGCUCCGCCAGCCAUCCAAUGCCAAACCGCUUAAAGCCAACGAAAAUUGCCAGCGACAA ......(((........((((((((((((..........))))))))))))......((((.(((.....)))................((.....))........))))...))).... ( -24.60) >consensus ACAUACCACCAGACAUAUUGCGUAUACGCUGACCAUAUUGCGUAUACGCAACCAACACAAUAGCUCCGCCAGCCAUCCAAUGCCAAACCGCUUAAAGCCA____AAAUUGCCAGCGACAA .................((((((((((((..........))))))))))))...............(((..((((.....)).......((.....))...........))..))).... (-20.30 = -20.70 + 0.40)

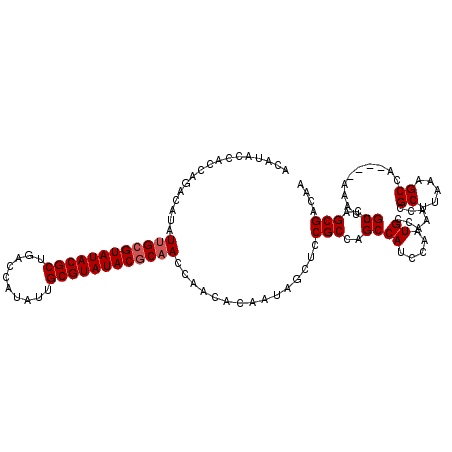
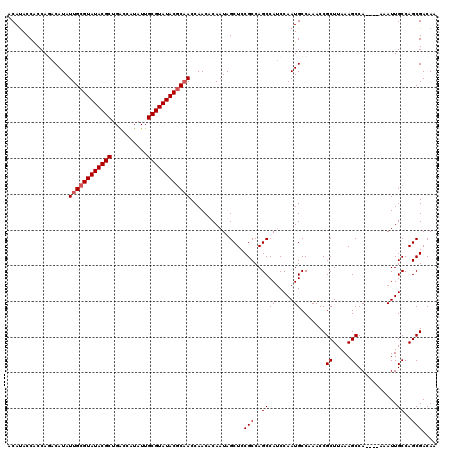
| Location | 21,962,053 – 21,962,169 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.27 |
| Mean single sequence MFE | -40.48 |
| Consensus MFE | -37.32 |
| Energy contribution | -37.92 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.92 |
| SVM decision value | 4.23 |
| SVM RNA-class probability | 0.999842 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21962053 116 - 22224390 UUGUCGCUGGCAAUUU----UGGCUUUAAGCGGUUUGGCAUUGGAUGGCUGGCGGAGCUAUUGUGUUGGUUGAGUAUACGCAAUAUGGUCAGCGUAUACGCAAUAUGUCUGGUGGUAUGU .(..(((..(((((..----(((((((.(((.((((......)))).)))...)))))))..))....((((.((((((((..........)))))))).)))).)).)..)))..)... ( -35.80) >DroSec_CAF1 24435 116 - 1 UUGUCGCUGGCAAUUU----UGGCUUUAAGCGGUUUGGCAUUGGAUGGCUGGCGGAGCUAUUGUGUUGGUUGCGUAUACGCAAUAUGGUCAGCGUAUACGCAAUAUGUCUGGUGGUAUGU .(..(((..(((((..----(((((((.(((.((((......)))).)))...)))))))..))....(((((((((((((..........))))))))))))).)).)..)))..)... ( -41.50) >DroSim_CAF1 13651 116 - 1 UUGUCGCUGGCAAUUU----UGGCUUUAAGCGGUUUGGCAUUGGAUGGCUGGCGGAGCUAUUGUGUUGGUUGCGUAUACGCAAUAUGGUCAGCGUAUACGCAAUAUGUCUGGUGGUAUGU .(..(((..(((((..----(((((((.(((.((((......)))).)))...)))))))..))....(((((((((((((..........))))))))))))).)).)..)))..)... ( -41.50) >DroEre_CAF1 9693 119 - 1 UUGUCGCUGGCAAUUUU-GUUGGCUUUAAGCGGUUUGGCAUUGGCUGGCUGGCGGAGCUAUUGUGUUGGUGGCGUAUACGCAAUAUGGCCAGCGUAUACGCAAUAUGUGUGGCGGUGUGU ..(((((..(((.....-..((((((...((.(.(..((....))..).).)).))))))...)))..)))))......(((.(((.((((((((((.....)))))).)))).)))))) ( -40.20) >DroYak_CAF1 16763 120 - 1 UUGUCGCUGGCAAUUUUCGUUGGCUUUAAGCGGUUUGGCAUUGGAUGGCUGGCGGAGCUAUUGUGUUGGUUGCGUAUACGCAAUAUGGCCAGCGUAUACGCAAUAUGUGUGGCGGUAUGU (((((((..(((((((((((..(((((((((......)).))))..)))..))))))..)))))....(((((((((((((..........)))))))))))))....)))))))..... ( -43.40) >consensus UUGUCGCUGGCAAUUU____UGGCUUUAAGCGGUUUGGCAUUGGAUGGCUGGCGGAGCUAUUGUGUUGGUUGCGUAUACGCAAUAUGGUCAGCGUAUACGCAAUAUGUCUGGUGGUAUGU .(..((((((((((......(((((((.(((.((((......)))).)))...)))))))....))))(((((((((((((..........)))))))))))))....))))))..)... (-37.32 = -37.92 + 0.60)

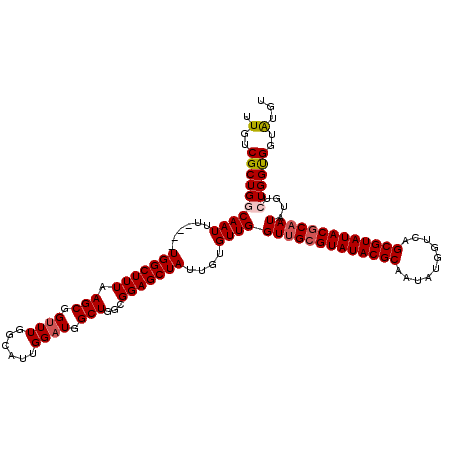
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:10:00 2006