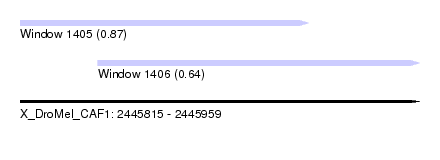
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,445,815 – 2,445,959 |
| Length | 144 |
| Max. P | 0.871583 |

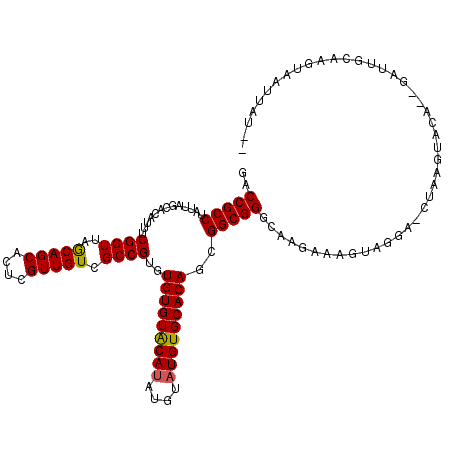
| Location | 2,445,815 – 2,445,919 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.15 |
| Mean single sequence MFE | -35.09 |
| Consensus MFE | -33.91 |
| Energy contribution | -33.36 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.07 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.871583 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2445815 104 + 22224390 GCCACCCCC------------CUCACUUUCGACGCCCUUGGACCGCCUAUUAGCACAUUUCGGUUAACAGCAGUUGUUGUCGCCGUGUGUGUACAUA----UGUGCACAGUGGCGGGCAA (((......------------.((......))((((.(((...(((.(((..((((((..((((..(((((....))))).)))).))))))..)))----.)))..))).))))))).. ( -33.30) >DroSec_CAF1 6655 120 + 1 GCCACCCCCUCACCCCACCCCCCCCUUUUCGACGCCCUUGGACCGCCUUUUAGCACAUUUCGGUUAGCAGCACUCGUUGUCGCCGUGUGUGUACAUAUGUAUGUGCACAGCGGCGGGCAA .................................((((...(((((...............))))).(((((....))))).(((((.(((((((((....)))))))))))))))))).. ( -35.96) >DroSim_CAF1 8846 117 + 1 GCCACCCCCUCACCC---ACCCCCCUUUUCGACGCCCUUGAACCGCCUAUUAGCACAUUUCGGUUAGCAGCACUCGUUGUCGCCGUGUGUGUGCAUAUGUAUGUGCACAGCGGCGGGCAA (((............---.........(((((.....))))).((((.....(((((...((((..(((((....))))).)))))))))(((((((....)))))))...))))))).. ( -36.00) >consensus GCCACCCCCUCACCC____CCCCCCUUUUCGACGCCCUUGGACCGCCUAUUAGCACAUUUCGGUUAGCAGCACUCGUUGUCGCCGUGUGUGUACAUAUGUAUGUGCACAGCGGCGGGCAA .................................((((...(((((...............))))).(((((....))))).(((((.(((((((((....)))))))))))))))))).. (-33.91 = -33.36 + -0.55)

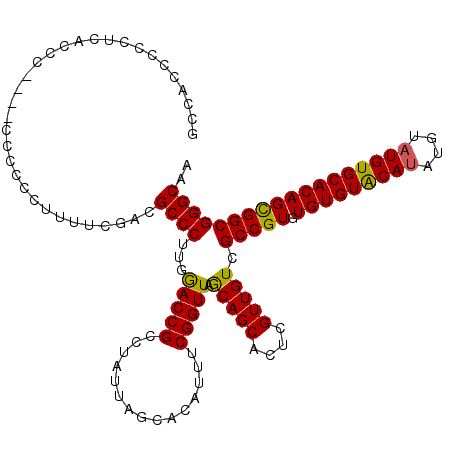
| Location | 2,445,843 – 2,445,959 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.35 |
| Mean single sequence MFE | -36.17 |
| Consensus MFE | -29.44 |
| Energy contribution | -29.33 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.643318 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2445843 116 + 22224390 GACCGCCUAUUAGCACAUUUCGGUUAACAGCAGUUGUUGUCGCCGUGUGUGUACAUA----UGUGCACAGUGGCGGGCAAGAAAGUAGGUCCUAAGUACAGAGAUUUCCAAUAAUUGCGA ..(((((.............((((..(((((....))))).))))..(((((((...----.)))))))..)))))((((......(((((((......)).))))).......)))).. ( -34.02) >DroSec_CAF1 6695 108 + 1 GACCGCCUUUUAGCACAUUUCGGUUAGCAGCACUCGUUGUCGCCGUGUGUGUACAUAUGUAUGUGCACAGCGGCGGGCAAGAAAGUAGGA-CUAAGU---------GCAAGUCAUUAU-- (((.((..(((((..(.((((.(((.(((((....)))))((((((.(((((((((....))))))))))))))))))..))))...)..-))))).---------))..))).....-- ( -36.10) >DroSim_CAF1 8883 117 + 1 AACCGCCUAUUAGCACAUUUCGGUUAGCAGCACUCGUUGUCGCCGUGUGUGUGCAUAUGUAUGUGCACAGCGGCGGGCAAGAAAGUAGGA-CUAAGUACACUGAUUGCGAGUAAUUAU-- ..(((((.....(((((...((((..(((((....))))).)))))))))(((((((....)))))))...)))))((((...(((.(..-.......))))..))))..........-- ( -38.40) >consensus GACCGCCUAUUAGCACAUUUCGGUUAGCAGCACUCGUUGUCGCCGUGUGUGUACAUAUGUAUGUGCACAGCGGCGGGCAAGAAAGUAGGA_CUAAGUACA__GAUUGCAAGUAAUUAU__ ..(((((.............((((..(((((....))))).))))..(((((((((....)))))))))..)))))............................................ (-29.44 = -29.33 + -0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:55:47 2006