
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 21,954,572 – 21,954,691 |
| Length | 119 |
| Max. P | 0.790993 |

| Location | 21,954,572 – 21,954,666 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.96 |
| Mean single sequence MFE | -30.98 |
| Consensus MFE | -19.88 |
| Energy contribution | -19.75 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.781367 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21954572 94 + 22224390 --------------------UGCAAUUGCCA-AAUUGCAUAUGCAAUUAAAUUGGGAAGAGCCAAAUGAGC-----CAGUUGGCGGAGCAACAAGGAUACCUGGCCAGGUGUGCGAGAAA --------------------(((((((....-)))))))..(((.......((((......))))....((-----(....)))...)))....(.((((((....)))))).)...... ( -26.00) >DroSec_CAF1 12917 119 + 1 CCAGGACUGCAACCAAAAAAAGAACUUGCCA-AAUUGCAUAUGCAAUCAAAUUGGGAAGAGCCAAAUGAGCCUUGCCACUUGGCGGUGUAAAAAGGAAGCCCGCCCAGGUGCGCGAGAAA ...((.......))..........(((.(((-((((((....)))))....)))).))).(((....).))(((((((((((((((.((.........))))).))))))).)))))... ( -34.40) >DroSim_CAF1 4611 119 + 1 CCAGGACUGCAACCAAAAAAAGAACUUGCCA-AAUUGCAUAUGCAAUCAAAUUGGGAAGAGCCAAAUGAGCCUUGCCACUUGGCGGUGCAAAAAGGAAGCCUGGCCAGGUGCGCGAGAAA ...((.......))..........(((.(((-((((((....)))))....)))).))).(((....).))(((((((((((((.(.((.........)).).)))))))).)))))... ( -34.80) >DroYak_CAF1 11279 116 + 1 CCAGGAGUUUAACCGAAAA----AAUUGCCAAAAUUGCAAAUGCUAUCAGACUGCGAAGAGCCAAAUGAGCCUUGCCAGUUGGCGGUGCAAAAAAGGAGCCCGUGCAGGUGCGCAAGAAA ...((.......)).....----..((((((...(((((...(((.......((((....(((((.((.((...)))).)))))..)))).......)))...))))).)).)))).... ( -28.74) >consensus CCAGGACUGCAACCAAAAA_AGAAAUUGCCA_AAUUGCAUAUGCAAUCAAAUUGGGAAGAGCCAAAUGAGCCUUGCCACUUGGCGGUGCAAAAAGGAAGCCCGGCCAGGUGCGCGAGAAA ........................(((((((..(((((....)))))....((((.....(((((.((........)).)))))((.((.........))))..)))).)).)))))... (-19.88 = -19.75 + -0.12)

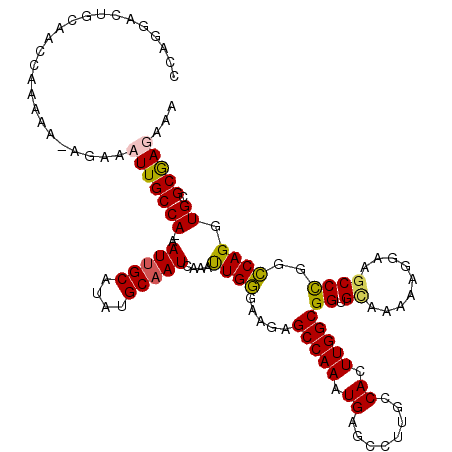
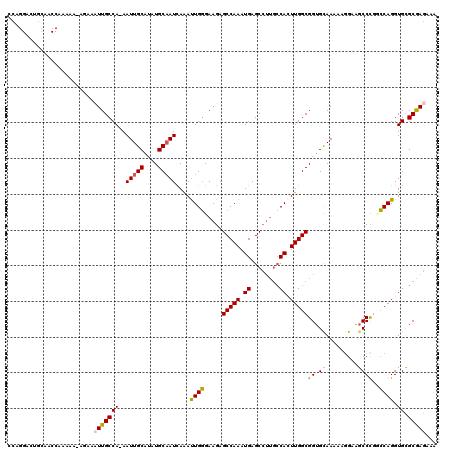
| Location | 21,954,591 – 21,954,691 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.68 |
| Mean single sequence MFE | -32.15 |
| Consensus MFE | -16.32 |
| Energy contribution | -16.40 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.790993 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21954591 100 + 22224390 AUGCAAUUAAAUUGGGAAGAGCCAAAUGAGC-----CAGUUGGCGGAGCAACAAGGAUACCUGGCCAGGUGUGCGAGAAAAUUCGUG-----------G--GAGAGUGGCAGAAAAGG-- ...........((((......))))....((-----((.((.(((((.......(.((((((....)))))).).......))))).-----------.--..)).))))........-- ( -24.64) >DroSec_CAF1 12956 105 + 1 AUGCAAUCAAAUUGGGAAGAGCCAAAUGAGCCUUGCCACUUGGCGGUGUAAAAAGGAAGCCCGCCCAGGUGCGCGAGAAAAUUCGUG-----------G--GAGAGUGGCAGAAAAGC-- ..((..(((..((((......)))).)))...((((((((((((((.((.........)))))))......((((((....))))))-----------.--..)))))))))....))-- ( -31.90) >DroSim_CAF1 4650 105 + 1 AUGCAAUCAAAUUGGGAAGAGCCAAAUGAGCCUUGCCACUUGGCGGUGCAAAAAGGAAGCCUGGCCAGGUGCGCGAGAAAACUCGUG-----------G--GAGAGUGGCAGAAAAGC-- .(((..((...((((......))))(((((.(((((((((((((.(.((.........)).).)))))))).)))))....))))).-----------.--..))...))).......-- ( -34.30) >DroEre_CAF1 4563 112 + 1 AUGCUAUCAAAUUUCGAAGGGCCAAAUGAGCCUUGCCAGUUGGCGGUGCAAAAAGGAAGCCCUGUGAGGUGCA--AGAGG----UGGCAGGAGAACCCGGAGGAAGUGGCAGAAAAGG-- .((((((....((((...(((.....((.((((((((....))).(..(....(((....))).....)..).--.))))----)..))......)))...)))))))))).......-- ( -30.90) >DroYak_CAF1 11315 118 + 1 AUGCUAUCAGACUGCGAAGAGCCAAAUGAGCCUUGCCAGUUGGCGGUGCAAAAAAGGAGCCCGUGCAGGUGCGCAAGAAAAUUUGGGCAGAAGCGGCAG--GGAAGUGGCAGAAAAGGGU .((((((....((((......((((((....(((((((.(((((((.((.........)))))).))).)).)))))...))))))((....)).))))--....))))))......... ( -39.00) >consensus AUGCAAUCAAAUUGGGAAGAGCCAAAUGAGCCUUGCCAGUUGGCGGUGCAAAAAGGAAGCCCGGCCAGGUGCGCGAGAAAAUUCGUG___________G__GAGAGUGGCAGAAAAGG__ ....................((((.....((.(((((....))))).)).........((((.....)).))..................................)))).......... (-16.32 = -16.40 + 0.08)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:09:54 2006