
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 21,954,399 – 21,954,515 |
| Length | 116 |
| Max. P | 0.917775 |

| Location | 21,954,399 – 21,954,515 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.82 |
| Mean single sequence MFE | -25.81 |
| Consensus MFE | -14.50 |
| Energy contribution | -17.70 |
| Covariance contribution | 3.20 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.917775 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
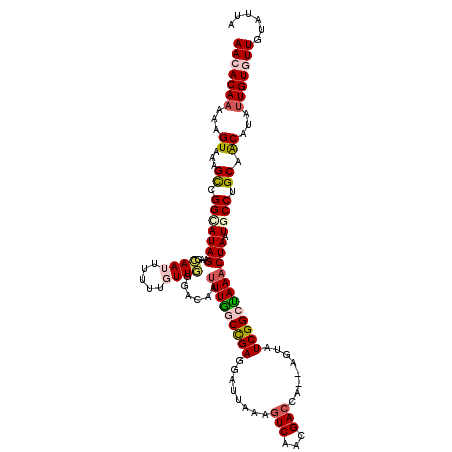
>X_DroMel_CAF1 21954399 116 + 22224390 UACUACAACACAACAUGCUGCAGGCAUUAGUUUAGCCGA--CU--UGGUCGUUGACUUUAAUCCUCGGCCAAAUAGUCCCAACAAAAAAUUGCUUCUAUACCGACUUUACAUUUUGUGUU ......(((((((.(((..((((((.........)))((--((--((((((..((......))..))))))...))))............))).((......)).....))).))))))) ( -25.20) >DroSec_CAF1 12667 98 + 1 UACUACAACACAAUAUGUGGCAGGCAUUAGUUUAGCCGAUAC----------------------UCGGCUAAAUUGUCCCAACAAAAAAUUGCUUCUAUGCCGGCUUUACUUUUUGUCUU .........((((...(((((.(((((.((((((((((....----------------------.))))))).((((....))))......)))...))))).)))..))...))))... ( -25.10) >DroSim_CAF1 4342 118 + 1 UAAUACAACACAAUAUGUUGCAGGCAUUAGUUUAGCCGAUACU--GGCUCGUUGACUUUAAUCCUCGGCUAAAUUGUCCCAACAAAAAAUUGCUUCUAUGCCGGCUUUACUUUUUGUCUU .....(((((.....))))).(((((..(((..(((((....(--(((.((..((......))..))))))....................((......)))))))..)))...))))). ( -26.20) >DroEre_CAF1 4253 119 + 1 UAAAACAACACAAAAUGUAGCAGGUAUUAGUUUACCCGAUACUUCUGGUCAUUGACUUUAAUCCUCAGCCAAAUUGUCGCAACAAAAA-UUCCAUCUAUGCCGGCUUUACUUUUUGUGUU ......(((((((((.((((.(((....((((.(((.((....)).)))....)))).....))))((((...((((....))))...-...((....))..)))).))).))))))))) ( -22.60) >DroYak_CAF1 11009 120 + 1 UAAUACAACACAUAAUGUAGCAGGUAAUAGGUUGCCCGAUACUUUUGGUCGUUGACUUUAAUCCUCAGCCAAAUUGACCGAACAAAAACUUCCAUCUAUGCCUGCUCUACUUUUUGUGUU ......((((((....((((((((((.(((((...........((((((((((((.........)))))......)))))))...........)))))))))))))..))....)))))) ( -29.95) >consensus UAAUACAACACAAAAUGUAGCAGGCAUUAGUUUAGCCGAUACU__UGGUCGUUGACUUUAAUCCUCGGCCAAAUUGUCCCAACAAAAAAUUGCUUCUAUGCCGGCUUUACUUUUUGUGUU ......(((((((...(((((.((((.((((((((((((.......((((...)))).......)))))))))(((......)))..........))))))).)))..))...))))))) (-14.50 = -17.70 + 3.20)



| Location | 21,954,399 – 21,954,515 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.82 |
| Mean single sequence MFE | -29.48 |
| Consensus MFE | -17.96 |
| Energy contribution | -19.23 |
| Covariance contribution | 1.27 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.810318 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21954399 116 - 22224390 AACACAAAAUGUAAAGUCGGUAUAGAAGCAAUUUUUUGUUGGGACUAUUUGGCCGAGGAUUAAAGUCAACGACCA--AG--UCGGCUAAACUAAUGCCUGCAGCAUGUUGUGUUGUAGUA (((((((.((((...((.(((((((.(((((....))))).......(((((((((.(((....)))..(.....--.)--))))))))))).))))).)).)))).)))))))...... ( -30.20) >DroSec_CAF1 12667 98 - 1 AAGACAAAAAGUAAAGCCGGCAUAGAAGCAAUUUUUUGUUGGGACAAUUUAGCCGA----------------------GUAUCGGCUAAACUAAUGCCUGCCACAUAUUGUGUUGUAGUA ...((((........((.(((((((.(((((....))))).......((((((((.----------------------....)))))))))).))))).))(((.....))))))).... ( -26.80) >DroSim_CAF1 4342 118 - 1 AAGACAAAAAGUAAAGCCGGCAUAGAAGCAAUUUUUUGUUGGGACAAUUUAGCCGAGGAUUAAAGUCAACGAGCC--AGUAUCGGCUAAACUAAUGCCUGCAACAUAUUGUGUUGUAUUA ..........(((...((((((.((((....)))).)))))).....(((((((((.(((....))).((.....--.)).)))))))))....))).((((((((...))))))))... ( -29.30) >DroEre_CAF1 4253 119 - 1 AACACAAAAAGUAAAGCCGGCAUAGAUGGAA-UUUUUGUUGCGACAAUUUGGCUGAGGAUUAAAGUCAAUGACCAGAAGUAUCGGGUAAACUAAUACCUGCUACAUUUUGUGUUGUUUUA (((((((((.........(((((.(((..((-(((((...((.(.....).)).)))))))...))).))).))....(((.((((((......)))))).))).)))))))))...... ( -29.20) >DroYak_CAF1 11009 120 - 1 AACACAAAAAGUAGAGCAGGCAUAGAUGGAAGUUUUUGUUCGGUCAAUUUGGCUGAGGAUUAAAGUCAACGACCAAAAGUAUCGGGCAACCUAUUACCUGCUACAUUAUGUGUUGUAUUA ((((((.((.((..((((((......(((..(((....(((((((.....)))))))(((....))))))..)))........((....)).....)))))))).)).))))))...... ( -31.90) >consensus AACACAAAAAGUAAAGCCGGCAUAGAAGCAAUUUUUUGUUGGGACAAUUUGGCCGAGGAUUAAAGUCAACGACCA__AGUAUCGGCUAAACUAAUGCCUGCAACAUAUUGUGUUGUAUUA (((((((...((...((.(((((((...((((.....))))......(((((((((........(((...)))........)))))))))))).)))).)).))...)))))))...... (-17.96 = -19.23 + 1.27)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:09:52 2006