
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 21,952,144 – 21,952,280 |
| Length | 136 |
| Max. P | 0.951739 |

| Location | 21,952,144 – 21,952,264 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.80 |
| Mean single sequence MFE | -30.36 |
| Consensus MFE | -27.02 |
| Energy contribution | -26.74 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.850727 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21952144 120 + 22224390 UUCAAAGUUAUGACAGUCGAACAGGCGUAGCGGUGAGUGCGUGUGCAUAAAAUCAGAUAUCAAAAAUUGUUUACAAAUAAUUUAAUAGCCGCAGCGCACGCACGUCACGCAUUCGUGUGC .(((..((((((...(.....)...))))))..)))((((((((((.........(....)..((((((((....))))))))..........))))))))))..(((((....))))). ( -34.00) >DroSec_CAF1 10600 114 + 1 UUCAAAGUUAUGACAGUCGAACAGGCGUAGCGGUGAGUGUGUGUGCAUAAAAUCAGAUAUCAAAAAUUGUUUACAAAUAAUUUAAUAGCCGCAGCGCACGCACGUCAC------GUGUGC ......((((((...(.....)...)))))).((((((((((((((.........(....)..((((((((....))))))))..........)))))))))).))))------...... ( -29.00) >DroSim_CAF1 2262 114 + 1 UUCAAAGUUAUGACAGUCGAACAGGCGUAGCGGUGAGUGUGUGUGCAUAAAAUCAGAUAUCAAAAAUUGUUUACAAAUAAUUUAAUAGCCGCAGCGCACGCACGUCAC------GUGUGC ......((((((...(.....)...)))))).((((((((((((((.........(....)..((((((((....))))))))..........)))))))))).))))------...... ( -29.00) >DroEre_CAF1 2191 120 + 1 UUCAAAGUUAUGACAGUCGAACAGGCGUAGCGGCGAGUGUGUGUGCAUAAAAUCAGAUAUCAAAAAUUGUUUACAAAUAAUUUAAUAGCCACAGCGCACGCACGUCACGCAUUCGUGUGC ............(((..((((...((((..(((((.(((((((.((.........(....)..((((((((....))))))))....)))))).))).))).))..)))).))))))).. ( -30.90) >DroYak_CAF1 10204 120 + 1 AUCGAAGUUAUGACAGCCGAACAGGCGAGGCAGUAAGAGUGUGUGCAUAAAAUCAGAUAUCAAAAAUUGUUUACAAAUAAUUUAAUAGCCACAGCACACGCACGUCACGCAUUCGUGUGC ..((((....((((.(((.....)))............((((((((.........(....)..((((((((....))))))))..........))))))))..))))....))))..... ( -28.90) >consensus UUCAAAGUUAUGACAGUCGAACAGGCGUAGCGGUGAGUGUGUGUGCAUAAAAUCAGAUAUCAAAAAUUGUUUACAAAUAAUUUAAUAGCCGCAGCGCACGCACGUCACGCAUUCGUGUGC ..........((((.(((.....)))..........((((((((((.........(....)..((((((((....))))))))..........))))))))))))))............. (-27.02 = -26.74 + -0.28)

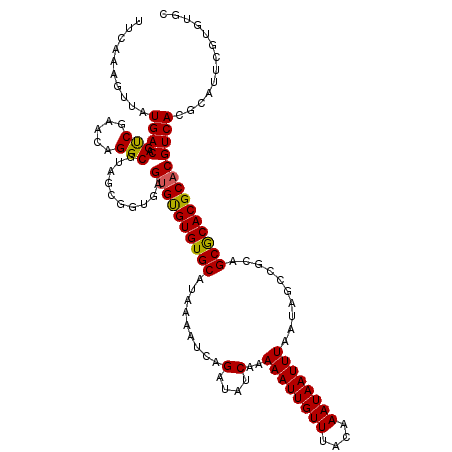

| Location | 21,952,184 – 21,952,280 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 91.62 |
| Mean single sequence MFE | -20.80 |
| Consensus MFE | -19.61 |
| Energy contribution | -19.42 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951739 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21952184 96 + 22224390 GUGUGCAUAAAAUCAGAUAUCAAAAAUUGUUUACAAAUAAUUUAAUAGCCGCAGCGCACGCACGUCACGCAUUCGUGUGCGCCGUAAAAACAGGAC-- ((((((.................((((((((....))))))))....((....)))))))).((.(((((....)))))))((.........))..-- ( -21.10) >DroSec_CAF1 10640 87 + 1 GUGUGCAUAAAAUCAGAUAUCAAAAAUUGUUUACAAAUAAUUUAAUAGCCGCAGCGCACGCACGUCAC------GUGUGCGCCGUAAAAACA---C-- (((.((.........(....)..((((((((....))))))))....))))).((((((((.......------))))))))..........---.-- ( -20.50) >DroSim_CAF1 2302 87 + 1 GUGUGCAUAAAAUCAGAUAUCAAAAAUUGUUUACAAAUAAUUUAAUAGCCGCAGCGCACGCACGUCAC------GUGUGCGCCGUAAAAACA---C-- (((.((.........(....)..((((((((....))))))))....))))).((((((((.......------))))))))..........---.-- ( -20.50) >DroEre_CAF1 2231 98 + 1 GUGUGCAUAAAAUCAGAUAUCAAAAAUUGUUUACAAAUAAUUUAAUAGCCACAGCGCACGCACGUCACGCAUUCGUGUGCGCCGCAAAAACAGGACUC ((((((.........(....)..((((((((....))))))))..........))))))((.((.(((((....)))))))..))............. ( -21.10) >consensus GUGUGCAUAAAAUCAGAUAUCAAAAAUUGUUUACAAAUAAUUUAAUAGCCGCAGCGCACGCACGUCAC______GUGUGCGCCGUAAAAACA___C__ (((.((.........(....)..((((((((....))))))))....))))).((((((((.............))))))))................ (-19.61 = -19.42 + -0.19)

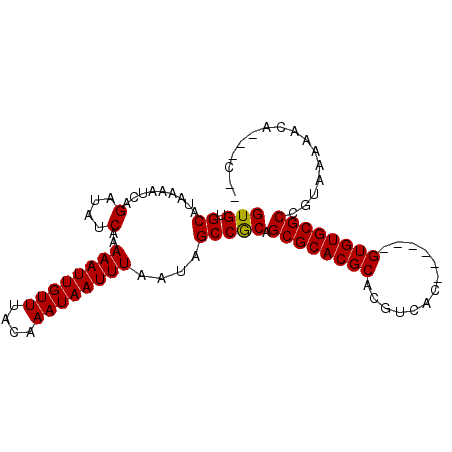
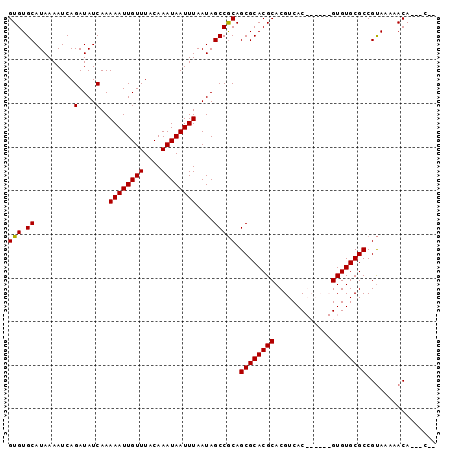
| Location | 21,952,184 – 21,952,280 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 91.62 |
| Mean single sequence MFE | -23.97 |
| Consensus MFE | -19.61 |
| Energy contribution | -19.55 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.604982 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21952184 96 - 22224390 --GUCCUGUUUUUACGGCGCACACGAAUGCGUGACGUGCGUGCGCUGCGGCUAUUAAAUUAUUUGUAAACAAUUUUUGAUAUCUGAUUUUAUGCACAC --((.((((....)))).)).((((....))))..(((((((.....(((.(((((((....(((....)))..))))))).)))....))))))).. ( -24.20) >DroSec_CAF1 10640 87 - 1 --G---UGUUUUUACGGCGCACAC------GUGACGUGCGUGCGCUGCGGCUAUUAAAUUAUUUGUAAACAAUUUUUGAUAUCUGAUUUUAUGCACAC --(---(((.....((((((((.(------(.....)).))))))))(((.(((((((....(((....)))..))))))).))).......)))).. ( -24.70) >DroSim_CAF1 2302 87 - 1 --G---UGUUUUUACGGCGCACAC------GUGACGUGCGUGCGCUGCGGCUAUUAAAUUAUUUGUAAACAAUUUUUGAUAUCUGAUUUUAUGCACAC --(---(((.....((((((((.(------(.....)).))))))))(((.(((((((....(((....)))..))))))).))).......)))).. ( -24.70) >DroEre_CAF1 2231 98 - 1 GAGUCCUGUUUUUGCGGCGCACACGAAUGCGUGACGUGCGUGCGCUGUGGCUAUUAAAUUAUUUGUAAACAAUUUUUGAUAUCUGAUUUUAUGCACAC ..((.((((....)))).)).((((....))))..(((((((.....(((.(((((((....(((....)))..))))))).)))....))))))).. ( -22.30) >consensus __G___UGUUUUUACGGCGCACAC______GUGACGUGCGUGCGCUGCGGCUAUUAAAUUAUUUGUAAACAAUUUUUGAUAUCUGAUUUUAUGCACAC ................((((.((((....))))..))))(((((...(((.(((((((....(((....)))..))))))).)))......))))).. (-19.61 = -19.55 + -0.06)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:09:48 2006