
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 21,951,222 – 21,951,506 |
| Length | 284 |
| Max. P | 0.999758 |

| Location | 21,951,222 – 21,951,320 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.63 |
| Mean single sequence MFE | -35.86 |
| Consensus MFE | -24.94 |
| Energy contribution | -25.82 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.570676 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21951222 98 - 22224390 CUGC--------------AAUCGGAGCCGUUUCUUUGGAGCUUGCGGCAAAGUUUGCAAACUCCCCCAG--------AGCCGCUGUUUAUCGGUUCUGUGGGCUUUGUUCAAAGUUCUGU ((..--------------...(((((((........((((.((((((......)))))).)))).((((--------(((((........)))))))).)))))))).....))...... ( -32.40) >DroSec_CAF1 9758 106 - 1 CUGC--------------AAUCGGAGCCGUUUCUUUGGAGUUUGCGGCAAAGUUUGCAAACUCCCCCAGCGCUGCAGCGCCGCUGUUUAUCGGUUCUGUGGGCUUUGUUCAAAGUUCUGU ..((--------------(..((((((((.......(((((((((((......)))))))))))..((((((......).))))).....)))))))).(((((((....)))))))))) ( -38.10) >DroSim_CAF1 1419 106 - 1 CUGC--------------AAUCGGAGCCGUUUCUUUGGAGUUUGCGGCAAAGUUUGCAAACUCCCCCAGCGCUGCAGCGCCGCUGUUUAUCGGUUCUGUGGGCUUUGUUCAAAGUUCUGU ..((--------------(..((((((((.......(((((((((((......)))))))))))..((((((......).))))).....)))))))).(((((((....)))))))))) ( -38.10) >DroEre_CAF1 1361 112 - 1 CUGCAAGCUGCAAGCAGCAAGCGGAGCCGUUUCUUUGAAGUUUGCGGCGAAGUUUGAAAACUCCCCCAG--------CGCCGCUGUUUAUCGGUUCUGUGGGCUUUGUUUAAAGUUCUGU ((((..((((....))))..)))).(((((..((....))...)))))(((.((((((.....((((((--------.((((........)))).))).))).....)))))).)))... ( -40.20) >DroYak_CAF1 9320 98 - 1 CUGC--------------AAGCGGAGCCGUUUCUUUGAAGUUUGCGGCAAAGUUUGCCAGCUCCCCCAG--------CGCCGCUGUUUAUCGGUUCUGUGGGCUUUGUUUAAAGUUCUGU ..((--------------((((...(((((..((....))...)))))...))))))(((..(((.(((--------.((((........)))).))).)))..)))............. ( -30.50) >consensus CUGC______________AAUCGGAGCCGUUUCUUUGGAGUUUGCGGCAAAGUUUGCAAACUCCCCCAG________CGCCGCUGUUUAUCGGUUCUGUGGGCUUUGUUCAAAGUUCUGU .....................((((((((.......(((((((((((......)))))))))))..(((.............))).....)))))))).(((((((....)))))))... (-24.94 = -25.82 + 0.88)


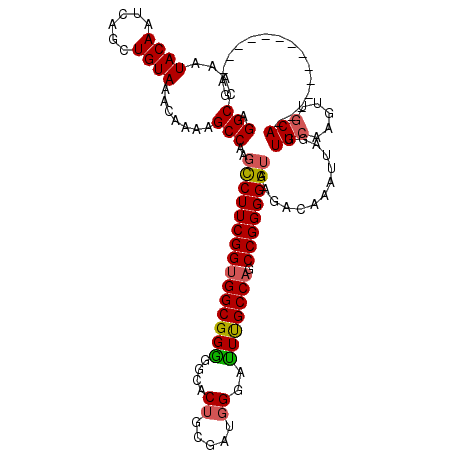
| Location | 21,951,320 – 21,951,426 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.87 |
| Mean single sequence MFE | -36.27 |
| Consensus MFE | -24.78 |
| Energy contribution | -24.86 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.548015 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21951320 106 - 22224390 AGGCCAAAUACAAUCAGCUGUAAACAAAAGCCAACCCUUCGGUGGCGGGGGCACUUCGAUGGGAUUCGCCAGCCGGGGGUAAGACAAAUUAGUUGCAAGUUGCA--------------CG .(((....((((......)))).......))).(((((.((((((((((....((.....))..))))))).))))))))...........((.((.....)))--------------). ( -28.70) >DroSec_CAF1 9864 106 - 1 AGGCCAAAUACAAUCAGCUGUAAACAAAAGCCAAGCCUUCGGUGGCGGGGGCACUGCGAUGGGACUUGCCAGCCGGGGGUAAGACAAAUUACUUGCAAGUUGCA--------------AG .(((....((((......)))).......)))..(((((((((((((((..(.(......))..))))))).))))))))...........(((((.....)))--------------)) ( -34.00) >DroSim_CAF1 1525 106 - 1 AGGCCAAAUACAAUCAGCUGUAAACAAAAGCCAAGCCUUCGGUGGCGGGGGCACUGCGAUGGGACUUGCCAGCCGGGGGUAAGACAAAUUACUUGCAAGUUGCA--------------AG .(((....((((......)))).......)))..(((((((((((((((..(.(......))..))))))).))))))))...........(((((.....)))--------------)) ( -34.00) >DroEre_CAF1 1473 118 - 1 AGGCCAAAUACAAUCAGCUGUAAACAAAAGCCAAGUCUUCGGUGGCGGGG--ACUCCGGUUGGAUUUGCCAGCCGGGGGUAAGACAAAUUAGUUGGAAGUUGCAAGUUGCAGGCUGCAAG .((((........(((((((.........(((.((((((((....)))))--)))((((((((.....)))))))).))).........)))))))....(((.....)))))))..... ( -41.77) >DroYak_CAF1 9418 120 - 1 AGGCCAAAUACAAUCAGCUGUAAACAAAAGCCAAGCCUUCGGAGGCGGAGGCACUUCGGUGGGAUUUGCCAGCCGGGGGUAAGACAAAUUAGUUGCAAGUUGCAAGUUGCAAGUUGCAAG .(((....((((......)))).......)))..((((((((.((((((..(((....)))...))))))..))))))))............((((((.((((.....)))).)))))). ( -42.90) >consensus AGGCCAAAUACAAUCAGCUGUAAACAAAAGCCAAGCCUUCGGUGGCGGGGGCACUGCGAUGGGAUUUGCCAGCCGGGGGUAAGACAAAUUAGUUGCAAGUUGCA______________AG .(((....((((......)))).......)))..(((((((((((((((....((.....))..))))))).)))))))).............(((.....)))................ (-24.78 = -24.86 + 0.08)


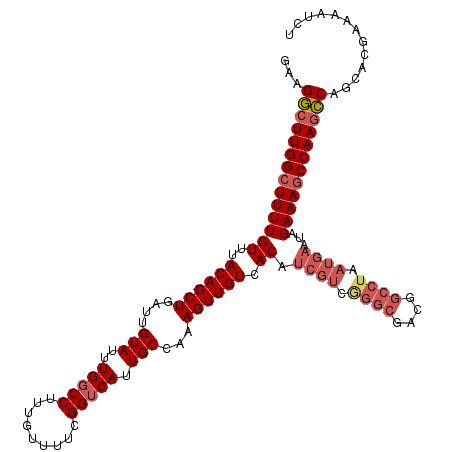
| Location | 21,951,386 – 21,951,506 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.25 |
| Mean single sequence MFE | -40.82 |
| Consensus MFE | -34.94 |
| Energy contribution | -36.66 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.27 |
| Structure conservation index | 0.86 |
| SVM decision value | 4.01 |
| SVM RNA-class probability | 0.999758 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21951386 120 + 22224390 GAAGGGUUGGCUUUUGUUUACAGCUGAUUGUAUUUGGCCUUUGUUUUCGGUCAUUGCCAAAGUUGUCACAUCGUCGGGCGAAGGCCUAAUGAAUAGAAAGCCAAGCCAGCACGAAAAUCU ...((.(((((((((((..((((((....(((..(((((.........))))).)))...)))))).)).((((..(((....)))..))))...))))))))).))............. ( -41.70) >DroSec_CAF1 9930 120 + 1 GAAGGCUUGGCUUUUGUUUACAGCUGAUUGUAUUUGGCCUUUGUUUUCGGUCAUUGCCAAAGUUGUCACAUCGUCAGGCGACGGCCUAAUGAAUAGAAAGCCAAGCCAGCACGAAAAUCU ...((((((((((((((..((((((....(((..(((((.........))))).)))...)))))).)).((((.((((....)))).))))...))))))))))))............. ( -43.00) >DroSim_CAF1 1591 120 + 1 GAAGGCUUGGCUUUUGUUUACAGCUGAUUGUAUUUGGCCUUUGUUUUCGGUCAUUGCCAAAGUUGUCACAUCGUCAGGCGACGGCCUAAUGAAUAGAAAGCCAAGCCAGCACGAAAAUCU ...((((((((((((((..((((((....(((..(((((.........))))).)))...)))))).)).((((.((((....)))).))))...))))))))))))............. ( -43.00) >DroEre_CAF1 1551 120 + 1 GAAGACUUGGCUUUUGUUUACAGCUGAUUGUAUUUGGCCUUUGUUUUCGGUCAUUGCAAAAGUUGUCACAUCGUCGGGCGGCGGCCUAAUGAAUUGAAAGCCAAGUCAACACGAAAAUCU ...((((((((((((((..((((((..(((((..(((((.........))))).))))).)))))).)).((((..(((....)))..))))...))))))))))))............. ( -44.30) >DroYak_CAF1 9498 107 + 1 GAAGGCUUGGCUUUUGUUUACAGCUGAUUGUAUUUGGCCUUUGUUUUCGGGCAUUGCAAAAGUUGUCACAUCGUCGGGCAAC-------------GAAAACCAAGCCAACACGAAAAUCU ...(((((((.(((((((.((((((..(((((....((((........))))..))))).))))))......(.....))))-------------)))).)))))))............. ( -32.10) >consensus GAAGGCUUGGCUUUUGUUUACAGCUGAUUGUAUUUGGCCUUUGUUUUCGGUCAUUGCCAAAGUUGUCACAUCGUCGGGCGACGGCCUAAUGAAUAGAAAGCCAAGCCAGCACGAAAAUCU ...((((((((((((((..((((((....(((..(((((.........))))).)))...)))))).)).((((.((((....)))).))))...))))))))))))............. (-34.94 = -36.66 + 1.72)


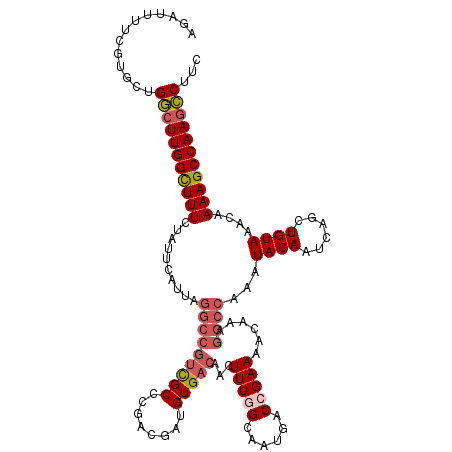
| Location | 21,951,386 – 21,951,506 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.25 |
| Mean single sequence MFE | -31.24 |
| Consensus MFE | -27.44 |
| Energy contribution | -28.60 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.980076 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21951386 120 - 22224390 AGAUUUUCGUGCUGGCUUGGCUUUCUAUUCAUUAGGCCUUCGCCCGACGAUGUGACAACUUUGGCAAUGACCGAAAACAAAGGCCAAAUACAAUCAGCUGUAAACAAAAGCCAACCCUUC .............((.((((((((..........(((((((((........))))....(((((......))))).....)))))...((((......))))....)))))))).))... ( -27.60) >DroSec_CAF1 9930 120 - 1 AGAUUUUCGUGCUGGCUUGGCUUUCUAUUCAUUAGGCCGUCGCCUGACGAUGUGACAACUUUGGCAAUGACCGAAAACAAAGGCCAAAUACAAUCAGCUGUAAACAAAAGCCAAGCCUUC .............(((((((((((..........(((((((((........)))))...(((((......)))))......))))...((((......))))....)))))))))))... ( -38.10) >DroSim_CAF1 1591 120 - 1 AGAUUUUCGUGCUGGCUUGGCUUUCUAUUCAUUAGGCCGUCGCCUGACGAUGUGACAACUUUGGCAAUGACCGAAAACAAAGGCCAAAUACAAUCAGCUGUAAACAAAAGCCAAGCCUUC .............(((((((((((..........(((((((((........)))))...(((((......)))))......))))...((((......))))....)))))))))))... ( -38.10) >DroEre_CAF1 1551 120 - 1 AGAUUUUCGUGUUGACUUGGCUUUCAAUUCAUUAGGCCGCCGCCCGACGAUGUGACAACUUUUGCAAUGACCGAAAACAAAGGCCAAAUACAAUCAGCUGUAAACAAAAGCCAAGUCUUC .............(((((((((((..........((((..((.....)).((..(......)..))...............))))...((((......))))....)))))))))))... ( -28.00) >DroYak_CAF1 9498 107 - 1 AGAUUUUCGUGUUGGCUUGGUUUUC-------------GUUGCCCGACGAUGUGACAACUUUUGCAAUGCCCGAAAACAAAGGCCAAAUACAAUCAGCUGUAAACAAAAGCCAAGCCUUC .............(((((((((((.-------------((((((((.((.((..(......)..)).))..))........)))....((((......))))))).)))))))))))... ( -24.40) >consensus AGAUUUUCGUGCUGGCUUGGCUUUCUAUUCAUUAGGCCGUCGCCCGACGAUGUGACAACUUUGGCAAUGACCGAAAACAAAGGCCAAAUACAAUCAGCUGUAAACAAAAGCCAAGCCUUC .............(((((((((((..........(((((((((........)))))...(((((......)))))......))))...((((......))))....)))))))))))... (-27.44 = -28.60 + 1.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:09:45 2006