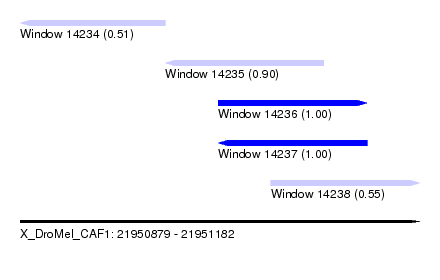
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 21,950,879 – 21,951,182 |
| Length | 303 |
| Max. P | 0.997958 |

| Location | 21,950,879 – 21,950,989 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.73 |
| Mean single sequence MFE | -22.93 |
| Consensus MFE | -17.36 |
| Energy contribution | -17.08 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.76 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.505737 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21950879 110 - 22224390 UGG--------CAACUUUUUCACCAUCUCCACUUUUCGACCACACAUCGGGGACAAACUUUACUUUGUAUUUGAGUGUGUG--UUUGCCAGUUGAUGAUUCUCGAAUUUCAUUUCUUUUU .(.--------...)........((((...(((...(((.(((((((((((.(((((......))))).))))).))))))--.)))..))).))))......(((......)))..... ( -20.20) >DroSec_CAF1 9407 111 - 1 UGG--------CAACUUU-UCCCCAUCUCUACUUUUCGGCCACACAUCGAGGACAAACUUUACUUUGUAUUUGAGUGUGUGUUUUUGCCAGUUGAUGAUUCUCGAAUUUCAUUCCUUUUU (((--------(((....-...((.............)).(((((((((((.(((((......))))).))))).))))))...))))))...(((((..........)))))....... ( -21.52) >DroSim_CAF1 1068 111 - 1 UGG--------CAACUUU-UCCCCAUCUCUACUUUUCGGCCACACAUCGAGGACAAACUUUACUUUGUAUUUGAGUGUGUGUGUUUGCCAGUUGAUGAUUCUCGAAUUUCAUUCCUUUUU (((--------(((....-...((.............)).(((((((((((.(((((......))))).))))).))))))...))))))...(((((..........)))))....... ( -21.52) >DroEre_CAF1 1018 109 - 1 CGG--------CAACUUUUUCCCCAUCUCCACUUUUCGACCACACAUCGAGGACAAACUUUACUUUGUAUUUGAGUGUGU--G-UUGCCAGUUGAUGAUUCUCGAAUUUCAUUUCUGUUU .(.--------...)........((((...(((...(((((((((.(((((.(((((......))))).)))))))))).--)-)))..))).))))......(((......)))..... ( -23.10) >DroYak_CAF1 8965 119 - 1 CGGCAACUCGGCAACUUUUUCCCCAUCUCCACUUUUCGACCACACAUCGAGGACAAACUUUACUUUGUAUCCGAGUGUGUGCG-UUGCCGGUUGAUGAUUCUCGAAUUUCAUUUAUAUUU (((((((..(....)......................(..(((((.(((.(((((((......))))).))))))))))..))-))))))((.(((((..........))))).)).... ( -28.30) >consensus UGG________CAACUUUUUCCCCAUCUCCACUUUUCGACCACACAUCGAGGACAAACUUUACUUUGUAUUUGAGUGUGUG_GUUUGCCAGUUGAUGAUUCUCGAAUUUCAUUUCUUUUU .......................((((...(((...(((.(((((((((((.(((((......))))).))))).))))))...)))..))).))))....................... (-17.36 = -17.08 + -0.28)



| Location | 21,950,989 – 21,951,109 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.17 |
| Mean single sequence MFE | -25.80 |
| Consensus MFE | -24.32 |
| Energy contribution | -24.16 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.895939 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21950989 120 - 22224390 ACUUUGGCAAUGCGCGAAUUAGCCAAAUAAUAAAUAAGUGCACGACAAAAGUGGCUAAGAAAUUUCGAACAACAAUGAAAUUACUCGGCUGCUCUUUUGAAAGACUUUUUGCCUAAUGCU .....((((((((((..((((......))))......)))))...((((((..((..((.(((((((........))))))).))..))..))..)))).........)))))....... ( -26.90) >DroSec_CAF1 9518 120 - 1 ACUUUGGCAAUGCGCGAAUUAGCCAAAUAAUAAAUAAGUGCACGACAAAAGUGGCUAAGAAAUUUCGAACAACAAUGAAAUUACUCGGCUGCUCUUUAGAAAGACUUUUUGCCUAAUGCC .....((((((((((..((((......))))......))))).......((..((..((.(((((((........))))))).))..))..))...............)))))....... ( -25.10) >DroSim_CAF1 1179 120 - 1 ACUUUGGCAAUGCGCGAAUUAGCCAAAUAAUAAAUAAGUGCACGACAAAAGUGGCUAAGAAAUUUCGAACAACAAUGAAAUUACUCGGCUGCUCUUUUGAAAGACUUUUUGCCUAAUGCC .....((((((((((..((((......))))......)))))...((((((..((..((.(((((((........))))))).))..))..))..)))).........)))))....... ( -26.90) >DroEre_CAF1 1127 120 - 1 ACUUUGGCAACGCGCGAAUUAGCCAAAUAAUAAAUAAGUGCACGACAAAAGUGGCUAAGAAAUUUCGAACAACAAUGAAAUUACUCGGCUGCUCUUUUGAAAGACUUUUUGCCUAAUGCU .....(((((.((((..((((......))))......))))....((((((..((..((.(((((((........))))))).))..))..))..)))).........)))))....... ( -26.40) >DroYak_CAF1 9084 120 - 1 ACUUUGGCAAUGCGCGAAUUAGCCAAAUAAUAAAUAAGUGCACGACAAAAGUGGCUAAGAAAUUUCGAACAACAAUGAAAUUACUCGGUUGCUCUUUUAAAAGACUUUUUGCCUAAUGCU .....((((((((((..((((......))))......))))).......((..((..((.(((((((........))))))).))..))..))...............)))))....... ( -23.70) >consensus ACUUUGGCAAUGCGCGAAUUAGCCAAAUAAUAAAUAAGUGCACGACAAAAGUGGCUAAGAAAUUUCGAACAACAAUGAAAUUACUCGGCUGCUCUUUUGAAAGACUUUUUGCCUAAUGCU .....(((((.((((..((((......))))......))))........((..((..((.(((((((........))))))).))..))..))...............)))))....... (-24.32 = -24.16 + -0.16)

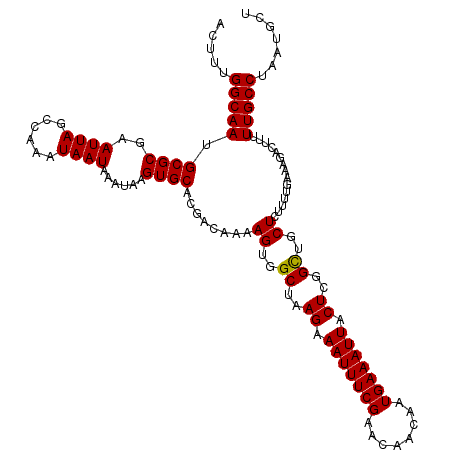
| Location | 21,951,029 – 21,951,142 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.80 |
| Mean single sequence MFE | -25.42 |
| Consensus MFE | -21.98 |
| Energy contribution | -21.82 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.56 |
| SVM RNA-class probability | 0.995331 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21951029 113 + 22224390 UUUCAUUGUUGUUCGAAAUUUCUUAGCCACUUUUGUCGUGCACUUAUUUAUUAUUUGGCUAAUUCGCGCAUUGCCAAAGUGGCUGC-------UUUUAUUUUUUUCUUUCCCAAGGAGAA .......................((((((((((.((.((((.(..((.....))..)((......)))))).)).)))))))))).-------.......((((((((....)))))))) ( -23.90) >DroSec_CAF1 9558 120 + 1 UUUCAUUGUUGUUCGAAAUUUCUUAGCCACUUUUGUCGUGCACUUAUUUAUUAUUUGGCUAAUUCGCGCAUUGCCAAAGUGGCUGCAUUUAUUUUUUAUUUUUUUCUUUCCCAAGGAGAA ..............(((((....((((((((((.((.((((.(..((.....))..)((......)))))).)).)))))))))).....))))).....((((((((....)))))))) ( -24.20) >DroSim_CAF1 1219 120 + 1 UUUCAUUGUUGUUCGAAAUUUCUUAGCCACUUUUGUCGUGCACUUAUUUAUUAUUUGGCUAAUUCGCGCAUUGCCAAAGUGGCUGCAUUUAUUUUUUAUUUUUUUCGUUCCCAAGGAGAA ((((.(((..(..(((((.....((((((((((.((.((((.(..((.....))..)((......)))))).)).))))))))))....((.....))....)))))..).))).)))). ( -25.30) >DroEre_CAF1 1167 114 + 1 UUUCAUUGUUGUUCGAAAUUUCUUAGCCACUUUUGUCGUGCACUUAUUUAUUAUUUGGCUAAUUCGCGCGUUGCCAAAGUGGCUGCAUUUA--UU----UUUUUGGCUUCCCAAGGAGAA .......................((((((((((.(((((((..(((.(((.....))).)))...)))))..)).))))))))))......--((----(((((((....))))))))). ( -26.80) >DroYak_CAF1 9124 116 + 1 UUUCAUUGUUGUUCGAAAUUUCUUAGCCACUUUUGUCGUGCACUUAUUUAUUAUUUGGCUAAUUCGCGCAUUGCCAAAGUGGCUGCAUUUGGUUU----UUUUUGCUUUGCCAAGGAGAA ..................(((((((((((((((.((.((((.(..((.....))..)((......)))))).)).)))))))))(((...(((..----.....))).)))..)))))). ( -26.90) >consensus UUUCAUUGUUGUUCGAAAUUUCUUAGCCACUUUUGUCGUGCACUUAUUUAUUAUUUGGCUAAUUCGCGCAUUGCCAAAGUGGCUGCAUUUA_UUUUUAUUUUUUUCUUUCCCAAGGAGAA ((((.(((......((....)).((((((((((.((.((((.(..((.....))..)((......)))))).)).))))))))))..........................))).)))). (-21.98 = -21.82 + -0.16)

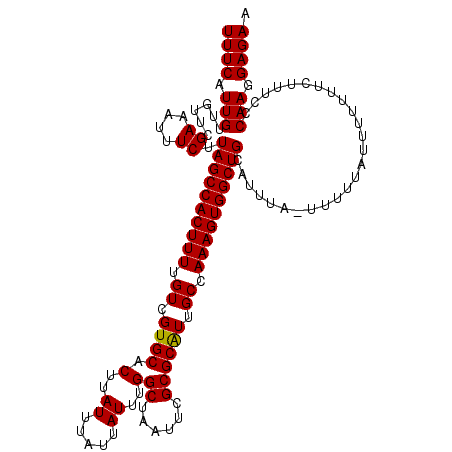

| Location | 21,951,029 – 21,951,142 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.80 |
| Mean single sequence MFE | -21.90 |
| Consensus MFE | -19.44 |
| Energy contribution | -19.64 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.97 |
| SVM RNA-class probability | 0.997958 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21951029 113 - 22224390 UUCUCCUUGGGAAAGAAAAAAAUAAAA-------GCAGCCACUUUGGCAAUGCGCGAAUUAGCCAAAUAAUAAAUAAGUGCACGACAAAAGUGGCUAAGAAAUUUCGAACAACAAUGAAA ((((.....))))..............-------..(((((((((.....(((((..((((......))))......))))).....)))))))))........................ ( -21.00) >DroSec_CAF1 9558 120 - 1 UUCUCCUUGGGAAAGAAAAAAAUAAAAAAUAAAUGCAGCCACUUUGGCAAUGCGCGAAUUAGCCAAAUAAUAAAUAAGUGCACGACAAAAGUGGCUAAGAAAUUUCGAACAACAAUGAAA ((((.....)))).......................(((((((((.....(((((..((((......))))......))))).....)))))))))........................ ( -21.00) >DroSim_CAF1 1219 120 - 1 UUCUCCUUGGGAACGAAAAAAAUAAAAAAUAAAUGCAGCCACUUUGGCAAUGCGCGAAUUAGCCAAAUAAUAAAUAAGUGCACGACAAAAGUGGCUAAGAAAUUUCGAACAACAAUGAAA ((((..(((....)))....................(((((((((.....(((((..((((......))))......))))).....))))))))).))))................... ( -21.60) >DroEre_CAF1 1167 114 - 1 UUCUCCUUGGGAAGCCAAAAA----AA--UAAAUGCAGCCACUUUGGCAACGCGCGAAUUAGCCAAAUAAUAAAUAAGUGCACGACAAAAGUGGCUAAGAAAUUUCGAACAACAAUGAAA ((((..((((....))))...----..--.......((((((((((....)((((..((((......))))......))))......))))))))).))))................... ( -23.80) >DroYak_CAF1 9124 116 - 1 UUCUCCUUGGCAAAGCAAAAA----AAACCAAAUGCAGCCACUUUGGCAAUGCGCGAAUUAGCCAAAUAAUAAAUAAGUGCACGACAAAAGUGGCUAAGAAAUUUCGAACAACAAUGAAA ...((.(((.....(((....----........)))(((((((((.....(((((..((((......))))......))))).....)))))))))................))).)).. ( -22.10) >consensus UUCUCCUUGGGAAAGAAAAAAAUAAAAA_UAAAUGCAGCCACUUUGGCAAUGCGCGAAUUAGCCAAAUAAUAAAUAAGUGCACGACAAAAGUGGCUAAGAAAUUUCGAACAACAAUGAAA ...((.(((...........................(((((((((.....(((((..((((......))))......))))).....)))))))))..((....))......))).)).. (-19.44 = -19.64 + 0.20)

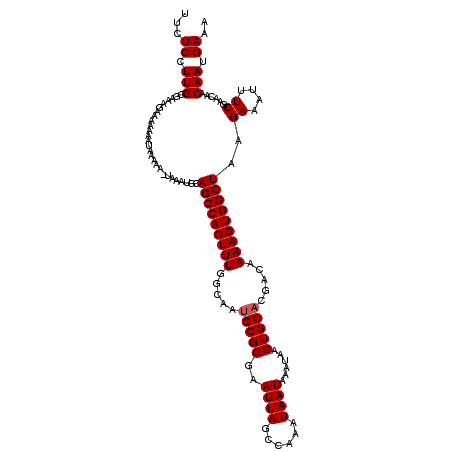
| Location | 21,951,069 – 21,951,182 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.80 |
| Mean single sequence MFE | -28.96 |
| Consensus MFE | -22.96 |
| Energy contribution | -25.24 |
| Covariance contribution | 2.28 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.551968 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21951069 113 + 22224390 CACUUAUUUAUUAUUUGGCUAAUUCGCGCAUUGCCAAAGUGGCUGC-------UUUUAUUUUUUUCUUUCCCAAGGAGAAAAGUAAUGAAAGUGCAAGGGAUAACUUUUUCGGUGGCCAA ..............(((((((....((((...(((.....)))...-------..(((((((((((((.....))))))))))))).....))))..(..(......)..)..))))))) ( -29.10) >DroSec_CAF1 9598 120 + 1 CACUUAUUUAUUAUUUGGCUAAUUCGCGCAUUGCCAAAGUGGCUGCAUUUAUUUUUUAUUUUUUUCUUUCCCAAGGAGAAAAGUAAUGAAAGUGCAAGGGAUAACUUUUUCGGUGGCCAA ..............(((((((....((((...(((.....)))........(((.(((((((((((((.....))))))))))))).))).))))..(..(......)..)..))))))) ( -29.90) >DroSim_CAF1 1259 120 + 1 CACUUAUUUAUUAUUUGGCUAAUUCGCGCAUUGCCAAAGUGGCUGCAUUUAUUUUUUAUUUUUUUCGUUCCCAAGGAGAAAAGUAAUGAAAGUGCAAGGGAUAACUUUUUCGGUGGCCAA ..............(((((((....((((...(((.....)))........(((.(((((((((((.(.....).))))))))))).))).))))..(..(......)..)..))))))) ( -28.20) >DroEre_CAF1 1207 114 + 1 CACUUAUUUAUUAUUUGGCUAAUUCGCGCGUUGCCAAAGUGGCUGCAUUUA--UU----UUUUUGGCUUCCCAAGGAGAAAAGUAAUGAAAGUGCAAGGGAUAACUUUUUCGGUGGCCAA ..............(((((((....((((((.(((.....))).))((((.--((----(((((((....))))))))).)))).......))))..(..(......)..)..))))))) ( -29.80) >DroYak_CAF1 9164 116 + 1 CACUUAUUUAUUAUUUGGCUAAUUCGCGCAUUGCCAAAGUGGCUGCAUUUGGUUU----UUUUUGCUUUGCCAAGGAGAAAAGUAAUGAAAGUGCAAGGGAUAACUUUUUCGGUGGCCAA ..............(((((((....((((...((((((((....)).))))))((----((.(((((((.(....)...))))))).))))))))..(..(......)..)..))))))) ( -27.80) >consensus CACUUAUUUAUUAUUUGGCUAAUUCGCGCAUUGCCAAAGUGGCUGCAUUUA_UUUUUAUUUUUUUCUUUCCCAAGGAGAAAAGUAAUGAAAGUGCAAGGGAUAACUUUUUCGGUGGCCAA ..............(((((((....((((...(((.....)))............(((((((((((((.....))))))))))))).....))))..(..(......)..)..))))))) (-22.96 = -25.24 + 2.28)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:09:42 2006