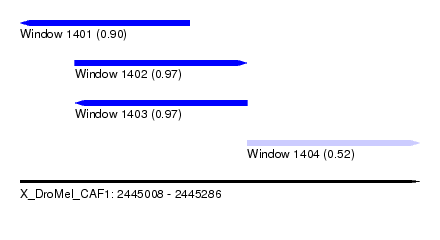
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,445,008 – 2,445,286 |
| Length | 278 |
| Max. P | 0.972866 |

| Location | 2,445,008 – 2,445,126 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.43 |
| Mean single sequence MFE | -32.90 |
| Consensus MFE | -31.47 |
| Energy contribution | -30.92 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.902342 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2445008 118 - 22224390 CACGCCGCUCUCUCGGCGUACUUUGCACAGGAAAAAAAAACGAAGGAAUUGCAGCAGAGGCGCUUGGAAAAGCUCAAGCUAACAGCGAUUUGUUUAAAAGGAAAAUCGAUUUUCCCAC-- .((((((......))))))......................(((.(((((((.((....))((((((......)))))).....))))))).)))....((((((....))))))...-- ( -30.90) >DroSec_CAF1 5841 119 - 1 CACGCCGCUCUUUCGGCGUACUUUGCACAG-AAAAAAAAGCAAAGGAAUUGCAGUAGAGGCGCUUGGAAAAGCCCAAACUAGCAGCGAUUUGUUUAAAAGGAAAAUCGAUUUUCCCACGA .((((((......)))))).((((((....-........))))))(((((((.((((.((.((((....))))))...)).)).)))))))........((((((....))))))..... ( -33.90) >DroSim_CAF1 8026 119 - 1 CACGCCGCUCUUUCGGCGUACUUUGCACAG-AAAAAAAAGCAAAGGAAUUGCAGUAGAGGCGCUUGGAAAAGCCCAAACUAGCAGCGAUUUGUUUAAAAGGAAAAUCGAUUUUCCCACGA .((((((......)))))).((((((....-........))))))(((((((.((((.((.((((....))))))...)).)).)))))))........((((((....))))))..... ( -33.90) >consensus CACGCCGCUCUUUCGGCGUACUUUGCACAG_AAAAAAAAGCAAAGGAAUUGCAGUAGAGGCGCUUGGAAAAGCCCAAACUAGCAGCGAUUUGUUUAAAAGGAAAAUCGAUUUUCCCACGA .((((((......)))))).((((((.............))))))((((((((((...((.((((....))))))..)))....)))))))........((((((....))))))..... (-31.47 = -30.92 + -0.55)

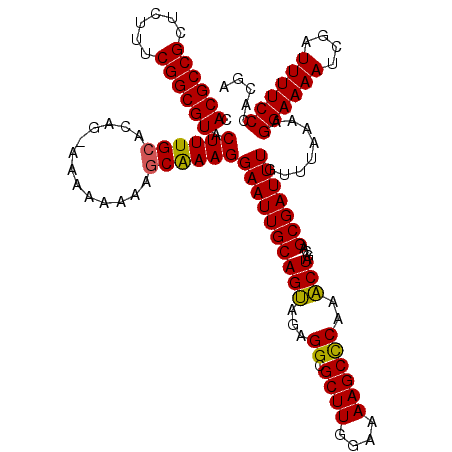
| Location | 2,445,046 – 2,445,166 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.55 |
| Mean single sequence MFE | -47.40 |
| Consensus MFE | -41.12 |
| Energy contribution | -40.69 |
| Covariance contribution | -0.43 |
| Combinations/Pair | 1.13 |
| Mean z-score | -3.55 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967660 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2445046 120 + 22224390 AGCUUGAGCUUUUCCAAGCGCCUCUGCUGCAAUUCCUUCGUUUUUUUUUCCUGUGCAAAGUACGCCGAGAGAGCGGCGUGAACCGGUGCAGAGAGAGAGAGAUUAAGUGAGAAGGGGUGA .(((((........)))))((((((.((.((....(((.((((((((((((((..(....(((((((......))))))).....)..))).))))))))))).))))))).)))))).. ( -46.40) >DroSec_CAF1 5881 115 + 1 AGUUUGGGCUUUUCCAAGCGCCUCUACUGCAAUUCCUUUGCUUUUUUUU-CUGUGCAAAGUACGCCGAAAGAGCGGCGUGAACCGGUGCAGAGAGAGAGCG----AGCGUGAAGAGGUGA .(((((((....)))))))((((((.((((.......(((((((((((.-(((..(....(((((((......))))))).....)..)))))))))))))----)))).).)))))).. ( -48.01) >DroSim_CAF1 8066 115 + 1 AGUUUGGGCUUUUCCAAGCGCCUCUACUGCAAUUCCUUUGCUUUUUUUU-CUGUGCAAAGUACGCCGAAAGAGCGGCGUGAACCGGUGCAGAGAGAGAGCG----AGUGUGAAGAGGUGA .(((((((....)))))))((((((.(.(((.....((((((((((((.-(((..(....(((((((......))))))).....)..)))))))))))))----)))))).)))))).. ( -47.80) >consensus AGUUUGGGCUUUUCCAAGCGCCUCUACUGCAAUUCCUUUGCUUUUUUUU_CUGUGCAAAGUACGCCGAAAGAGCGGCGUGAACCGGUGCAGAGAGAGAGCG____AGUGUGAAGAGGUGA .(((((((....)))))))((((((.((((........((((((((((..(((..(....(((((((......))))))).....)..))))))))))))).....))).).)))))).. (-41.12 = -40.69 + -0.43)

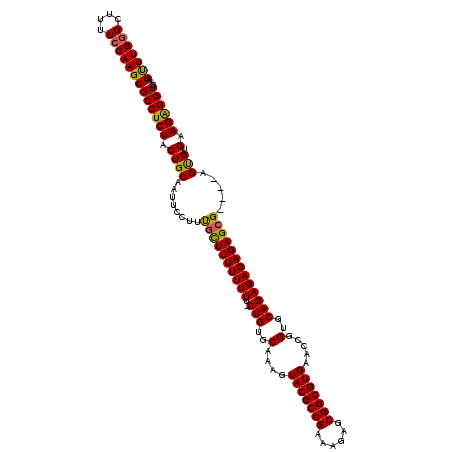
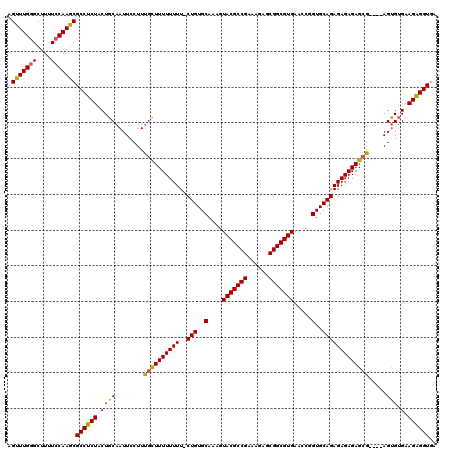
| Location | 2,445,046 – 2,445,166 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.55 |
| Mean single sequence MFE | -35.62 |
| Consensus MFE | -32.89 |
| Energy contribution | -32.79 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.972866 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2445046 120 - 22224390 UCACCCCUUCUCACUUAAUCUCUCUCUCUCUGCACCGGUUCACGCCGCUCUCUCGGCGUACUUUGCACAGGAAAAAAAAACGAAGGAAUUGCAGCAGAGGCGCUUGGAAAAGCUCAAGCU ..........................(((((((..((((((((((((......)))))).(((((...............)))))))))))..))))))).((((((......)))))). ( -33.26) >DroSec_CAF1 5881 115 - 1 UCACCUCUUCACGCU----CGCUCUCUCUCUGCACCGGUUCACGCCGCUCUUUCGGCGUACUUUGCACAG-AAAAAAAAGCAAAGGAAUUGCAGUAGAGGCGCUUGGAAAAGCCCAAACU ...............----.((.((((..(((((...((((((((((......)))))).((((((....-........))))))))))))))).))))))((((....))))....... ( -36.80) >DroSim_CAF1 8066 115 - 1 UCACCUCUUCACACU----CGCUCUCUCUCUGCACCGGUUCACGCCGCUCUUUCGGCGUACUUUGCACAG-AAAAAAAAGCAAAGGAAUUGCAGUAGAGGCGCUUGGAAAAGCCCAAACU ....((.(((.((..----(((.((((..(((((...((((((((((......)))))).((((((....-........))))))))))))))).)))))))..))))).))........ ( -36.80) >consensus UCACCUCUUCACACU____CGCUCUCUCUCUGCACCGGUUCACGCCGCUCUUUCGGCGUACUUUGCACAG_AAAAAAAAGCAAAGGAAUUGCAGUAGAGGCGCUUGGAAAAGCCCAAACU ..........................(((((((..((((((((((((......)))))).((((((.............))))))))))))..))))))).((((....))))....... (-32.89 = -32.79 + -0.11)

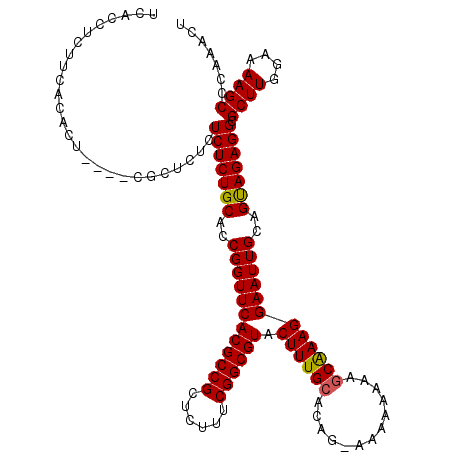
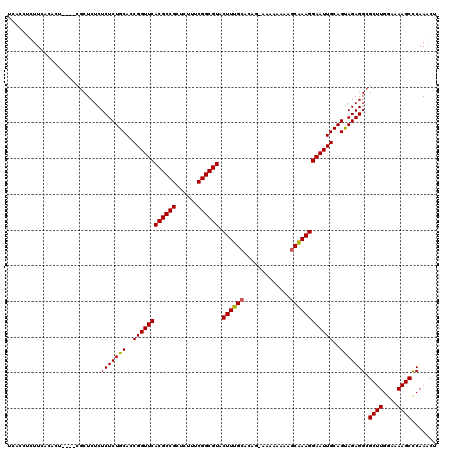
| Location | 2,445,166 – 2,445,286 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.08 |
| Mean single sequence MFE | -23.65 |
| Consensus MFE | -20.90 |
| Energy contribution | -21.23 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.88 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.522550 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2445166 120 + 22224390 GAGAGUGGAAGAAGGUGUUCAUUGGCAGAAAAUUAGCAAGGCGACAAACGUUUAUAAAUAUAAACACAUAUAUUCCAAUAUAAAAAUCGCAAAUAUUAAUGACGAGACAAGAGCGCCUGC ............((((((((.(((.......(((((....((((.....((((((....))))))...(((((....)))))....)))).....))))).......))))))))))).. ( -23.94) >DroSec_CAF1 5996 117 + 1 GAGAGAG---GAAGGUGUUCAUCGGCGGAAAAUUAGCAAGGCAACAAACGUUUAUAAAUAUAAACACAUAUAUUCCAAUAUAAAAAUCGCAAAUAUUAAUGACGAGACAAGAGCGCCUGC .......---..((((((((......((((.........(....)....((((((....)))))).......))))..........(((((........)).))).....)))))))).. ( -23.50) >DroSim_CAF1 8181 117 + 1 GAGAGAG---GAAGGUGUUCAUCGGCGGAAAAUUAGCAAGGCAACAAACGUUUAUAAAUAUAAACACAUAUAUUCCAAUAUAAAAAUCGCAAAUAUUAAUGACGAGACAAGAGCGCCUGC .......---..((((((((......((((.........(....)....((((((....)))))).......))))..........(((((........)).))).....)))))))).. ( -23.50) >consensus GAGAGAG___GAAGGUGUUCAUCGGCGGAAAAUUAGCAAGGCAACAAACGUUUAUAAAUAUAAACACAUAUAUUCCAAUAUAAAAAUCGCAAAUAUUAAUGACGAGACAAGAGCGCCUGC ............((((((((......((((.........(....)....((((((....)))))).......))))..........(((((........)).))).....)))))))).. (-20.90 = -21.23 + 0.33)

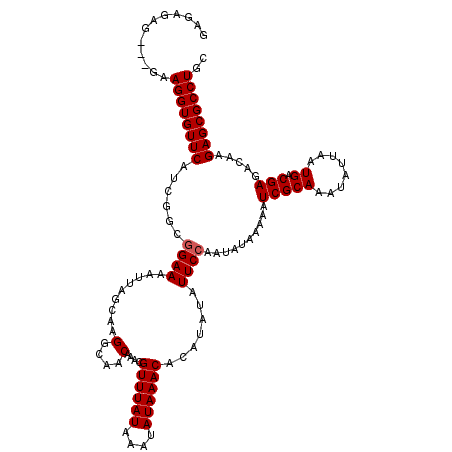
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:55:45 2006