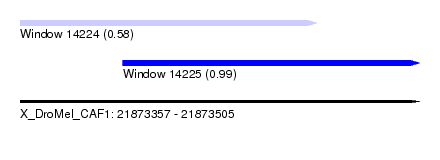
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 21,873,357 – 21,873,505 |
| Length | 148 |
| Max. P | 0.988667 |

| Location | 21,873,357 – 21,873,467 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 82.76 |
| Mean single sequence MFE | -34.39 |
| Consensus MFE | -19.46 |
| Energy contribution | -19.80 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.577423 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21873357 110 + 22224390 CCCAUAAGUAUGCUUACUUAUGUUCCCUAAGUUG-GGGCAUAAUGCUGUUGGUACUCCAUA--------GCAAUGGAUUCCCCAAUGCAGUGGGAAAAUAAAACGGCCACCUUCAGUCA ..((((((((....))))))))(((((...((((-(((.((..((((((.((....)))))--------)))....)).))))))).....)))))........(((........))). ( -35.50) >DroSec_CAF1 64371 110 + 1 CCCAUAAGUAUGCUUACUUAUGUUCCCUAAGUUG-GGGCAUAAUGCUGUUGGUACUCCACA--------GCAAUGGAUUCCCCAAUGCAUUGCGAAAAUAUGACGUUCACCGUGAGUCA ..((((((((....))))))))(((.(...((((-(((.((..((((((.((....)))))--------)))....)).))))))).....).)))....((((...........)))) ( -31.00) >DroEre_CAF1 36441 119 + 1 CCCAUAAGUAUGCUUACUUAAAUGCCCUAGGUUCAGGGCAUCAUGCUGUCGGUACUCCGGAACUCCGCUGCAAUGGAUUCCCCAAUGCAUUGCGAAAAUAUUACGGUCAUCUGCAGCCA ....((((((....)))))).(((((((......)))))))...((((.(((....(((......(((((((.(((.....))).))))..))).........)))....))))))).. ( -36.66) >consensus CCCAUAAGUAUGCUUACUUAUGUUCCCUAAGUUG_GGGCAUAAUGCUGUUGGUACUCCACA________GCAAUGGAUUCCCCAAUGCAUUGCGAAAAUAUAACGGUCACCUUCAGUCA ..((((((((....))))))))..(((........)))......((((((((....)))..........(((.(((.....))).)))..............)))))............ (-19.46 = -19.80 + 0.34)

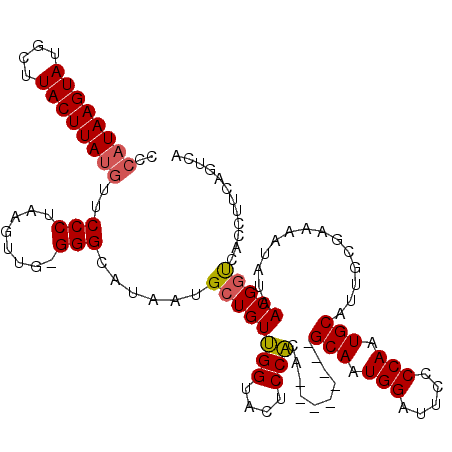

| Location | 21,873,395 – 21,873,505 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.86 |
| Mean single sequence MFE | -28.69 |
| Consensus MFE | -16.17 |
| Energy contribution | -16.40 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.56 |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.988667 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21873395 110 + 22224390 AUAAUGCUGUUGGUACUCCAUA--------GCAAUGGAUUCCCCAAUGCAGUGGGAAAAUAAAACGGCCACCUUCAGUCA--UUUGGCUUGAAAUUAAAUGACCGACAGAAUGAAAACAC ......((((((((..(((((.--------(((.(((.....))).))).)))))..........(((((..........--..)))))............))))))))........... ( -28.90) >DroSec_CAF1 64409 112 + 1 AUAAUGCUGUUGGUACUCCACA--------GCAAUGGAUUCCCCAAUGCAUUGCGAAAAUAUGACGUUCACCGUGAGUCAGCUUUAGCUUGAAAUUAAAAGACCGACAGAAUAAAAAUAU ......((((((((.(((.((.--------((((((.(((....))).))))))(((.........)))...)))))(((((....)).))).........))))))))........... ( -26.40) >DroEre_CAF1 36480 107 + 1 AUCAUGCUGUCGGUACUCCGGAACUCCGCUGCAAUGGAUUCCCCAAUGCAUUGCGAAAAUAUUACGGUCAUCUGCAGCCA--U-----------UUAAAAGACCUUUGGAGUAAAAGCAU ...(((((.....(((((((((....(((((((.(((.....))).))))..)))..........((((...........--.-----------......)))))))))))))..))))) ( -30.77) >consensus AUAAUGCUGUUGGUACUCCACA________GCAAUGGAUUCCCCAAUGCAUUGCGAAAAUAUAACGGUCACCUUCAGUCA__UUU_GCUUGAAAUUAAAAGACCGACAGAAUAAAAACAU ......((((((((................(((.(((.....))).)))................(((........)))......................))))))))........... (-16.17 = -16.40 + 0.23)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:09:31 2006