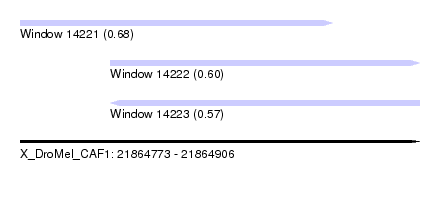
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 21,864,773 – 21,864,906 |
| Length | 133 |
| Max. P | 0.681161 |

| Location | 21,864,773 – 21,864,877 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.77 |
| Mean single sequence MFE | -32.45 |
| Consensus MFE | -27.85 |
| Energy contribution | -26.91 |
| Covariance contribution | -0.94 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.681161 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21864773 104 + 22224390 ----------UCUCUGCGAAGAGGCAAUUCAUUAGCUUUAAUUUAGGUAAAUGUCCUUGGUCCUUGUGGCG------UUGCCUCUUGCGGUGAAUGUCGCAAAUGCAACUAAAAUGUUGC ----------((.((((.((((((((((.((((.((((......)))).))))......(.((....))))------))))))))))))).))...........(((((......))))) ( -32.00) >DroVir_CAF1 27423 108 + 1 CUUG------CCAACGCGAAGAGGCAAUUCAUUAGCUUUAAUUUAGGUAAAUGUCCUUGGUCCUUGUGGUG------CUGCCUCUUGUGGCGAACGUUGCAAAUGCAACUAAAAUGUUGC .(((------(((.....(((((((....((((.((((......)))).)))).....(((((....)).)------))))))))).))))))..(((((....)))))........... ( -31.30) >DroGri_CAF1 25041 108 + 1 CUUG------CAAAUGCGAAGAGGCAAUUCAUUAGCUUUAAUUUAGGUAAAUGUCCUUGGUGCUUCUGGCG------UUGCCUCUCGUGGCGAACGUUGCAAAUGCAACUAAAAUGUUGC .(((------(((..((..(((((((...((((.((((......)))).)))).......))))))).))(------(((((......))).))).))))))..(((((......))))) ( -33.40) >DroWil_CAF1 26683 120 + 1 CAUACCAGCUCCAGUGCGAAGAGGCAAUUCAUUAGCUUUAAUUUAGGUAAAUGUCCUUGGUCCUUGUGGCGACGGCAUUGCCUCUUGUGGCGAACGUCGCAAAUGCAACUAAAAUGUUGC ..(((.((..(((((((......)))...((((.((((......)))).))))...))))..)).)))((((((...(((((......))))).))))))....(((((......))))) ( -35.50) >DroMoj_CAF1 35102 108 + 1 CUUG------CCAACGCGAAGAGGCAAUUCAUUAGCUUUAAUUUAGGUAAAUGUCCUUGGUCCUUGUGGUG------CUGCCUCUUGUGGCGAACGUUGCAAAUGCAACUAAAAUGUUGC .(((------(((.....(((((((....((((.((((......)))).)))).....(((((....)).)------))))))))).))))))..(((((....)))))........... ( -31.30) >DroAna_CAF1 20895 104 + 1 ----------ACUGUGCGAAGAGGCAAUUCAUUAGCUUUAAUUUAGGUAAAUGUCCUUGGUCCUUGUGGCG------UUGCCUCUUGCGGUGAGCGUCGCAAAUGCAACUAAAAUGUUGC ----------....((((((((((((((.((((.((((......)))).))))......(.((....))))------)))))))).((.....)).)))))...(((((......))))) ( -31.20) >consensus CUUG______CCAAUGCGAAGAGGCAAUUCAUUAGCUUUAAUUUAGGUAAAUGUCCUUGGUCCUUGUGGCG______UUGCCUCUUGUGGCGAACGUCGCAAAUGCAACUAAAAUGUUGC ..............(((((.(((((((.......(((...((..(((.......)))..))......))).......))))))))))))((((...))))....(((((......))))) (-27.85 = -26.91 + -0.94)

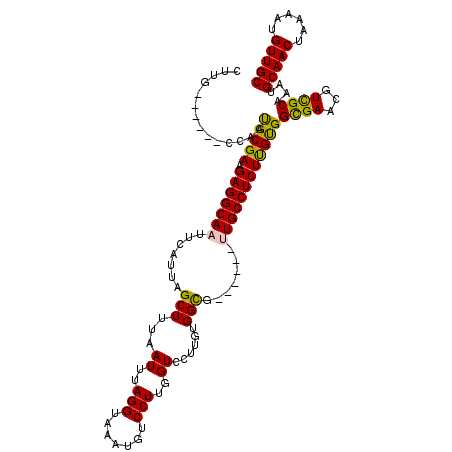
| Location | 21,864,803 – 21,864,906 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.30 |
| Mean single sequence MFE | -33.18 |
| Consensus MFE | -21.53 |
| Energy contribution | -21.37 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.600161 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21864803 103 + 22224390 AUUUAGGUAAAUGUCCUUGGUCCUUGUGGCG------UUGCCUCUUGCGGUGAAUGUCGCAAAUGCAACUAAAAUGUUGCGGUUACCGUCAUUUCC---------GCCCACA-GCCACG- .....((.(((((....((((((((((((((------(((((......))).))))))))))..(((((......)))))))..)))).)))))))---------.......-......- ( -30.90) >DroVir_CAF1 27457 105 + 1 AUUUAGGUAAAUGUCCUUGGUCCUUGUGGUG------CUGCCUCUUGUGGCGAACGUUGCAAAUGCAACUAAAAUGUUGCGGUUACCUCCAUUUUC---------AUUUGCUUACCACAU ...((((((((((....(((.......((((------..(((..(((..((....))..)))..(((((......)))))))))))).)))....)---------)))))))))...... ( -31.10) >DroGri_CAF1 25075 105 + 1 AUUUAGGUAAAUGUCCUUGGUGCUUCUGGCG------UUGCCUCUCGUGGCGAACGUUGCAAAUGCAACUAAAAUGUUGCGGUUACCGCUGUUUUC---------AUUUGCUUGCCACAU ....((((((.((((...((....)).))))------))))))...(((((((.((((((....)))))..(((((..(((((....)))))...)---------))))).))))))).. ( -32.50) >DroWil_CAF1 26723 119 + 1 AUUUAGGUAAAUGUCCUUGGUCCUUGUGGCGACGGCAUUGCCUCUUGUGGCGAACGUCGCAAAUGCAACUAAAAUGUUGCGGUAACCGCCAUUUUCAUUUGGGCCACCCGCU-UCCAACU .....((.....((...((((((..(((((((((...(((((......))))).))))))....(((((......)))))(((....))).....)))..))))))...)).-.)).... ( -41.90) >DroMoj_CAF1 35136 105 + 1 AUUUAGGUAAAUGUCCUUGGUCCUUGUGGUG------CUGCCUCUUGUGGCGAACGUUGCAAAUGCAACUAAAAUGUUGCGGUUACCUCCAUUUUC---------AUUUGCUUACCACAU ...((((((((((....(((.......((((------..(((..(((..((....))..)))..(((((......)))))))))))).)))....)---------)))))))))...... ( -31.10) >DroAna_CAF1 20925 104 + 1 AUUUAGGUAAAUGUCCUUGGUCCUUGUGGCG------UUGCCUCUUGCGGUGAGCGUCGCAAAUGCAACUAAAAUGUUGCGGUUACCAUCAUUCCC---------GCCCACU-GCCACAU .....((((((((....((((((((((((((------(((((......))).))))))))))..(((((......)))))))..)))).))))...---------......)-))).... ( -31.60) >consensus AUUUAGGUAAAUGUCCUUGGUCCUUGUGGCG______UUGCCUCUUGUGGCGAACGUCGCAAAUGCAACUAAAAUGUUGCGGUUACCGCCAUUUUC_________ACCCGCU_GCCACAU .....(((((.............((((((((......(((((......))))).)))))))).((((((......)))))).)))))................................. (-21.53 = -21.37 + -0.16)

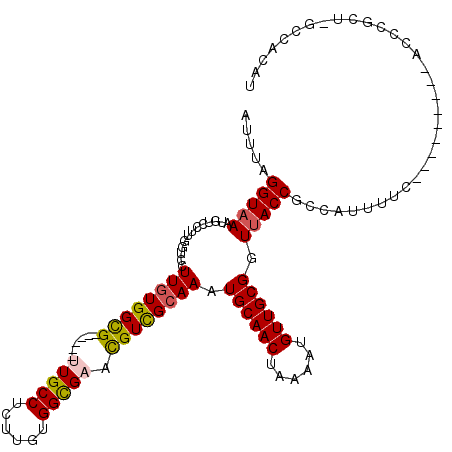
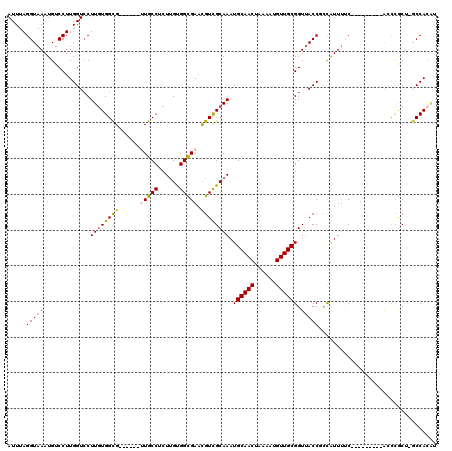
| Location | 21,864,803 – 21,864,906 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.30 |
| Mean single sequence MFE | -31.12 |
| Consensus MFE | -20.41 |
| Energy contribution | -19.93 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.565184 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
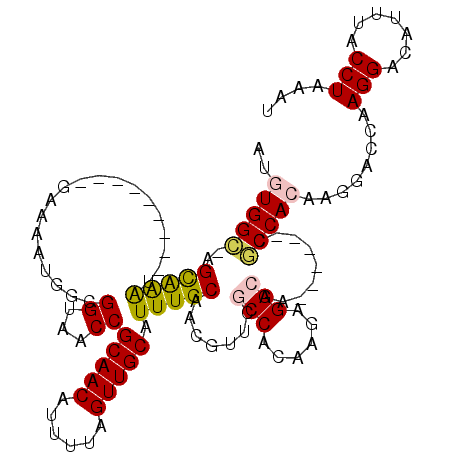
>X_DroMel_CAF1 21864803 103 - 22224390 -CGUGGC-UGUGGGC---------GGAAAUGACGGUAACCGCAACAUUUUAGUUGCAUUUGCGACAUUCACCGCAAGAGGCAA------CGCCACAAGGACCAAGGACAUUUACCUAAAU -.(((((-(((..((---------((............)))).))).....(((((.((((((........))))))..))))------))))))........(((.......))).... ( -31.20) >DroVir_CAF1 27457 105 - 1 AUGUGGUAAGCAAAU---------GAAAAUGGAGGUAACCGCAACAUUUUAGUUGCAUUUGCAACGUUCGCCACAAGAGGCAG------CACCACAAGGACCAAGGACAUUUACCUAAAU .((((((..((((((---------......((......))(((((......)))))))))))...(((.(((......)))))------))))))).......(((.......))).... ( -29.30) >DroGri_CAF1 25075 105 - 1 AUGUGGCAAGCAAAU---------GAAAACAGCGGUAACCGCAACAUUUUAGUUGCAUUUGCAACGUUCGCCACGAGAGGCAA------CGCCAGAAGCACCAAGGACAUUUACCUAAAU .(.((((..((((((---------(......((((...))))(((......))).)))))))...(((.(((......)))))------))))).).......(((.......))).... ( -28.60) >DroWil_CAF1 26723 119 - 1 AGUUGGA-AGCGGGUGGCCCAAAUGAAAAUGGCGGUUACCGCAACAUUUUAGUUGCAUUUGCGACGUUCGCCACAAGAGGCAAUGCCGUCGCCACAAGGACCAAGGACAUUUACCUAAAU ...(((.-....(((((((...((....))...)))))))(((((......)))))....((((((...(((......))).....))))))........)))(((.......))).... ( -34.70) >DroMoj_CAF1 35136 105 - 1 AUGUGGUAAGCAAAU---------GAAAAUGGAGGUAACCGCAACAUUUUAGUUGCAUUUGCAACGUUCGCCACAAGAGGCAG------CACCACAAGGACCAAGGACAUUUACCUAAAU .((((((..((((((---------......((......))(((((......)))))))))))...(((.(((......)))))------))))))).......(((.......))).... ( -29.30) >DroAna_CAF1 20925 104 - 1 AUGUGGC-AGUGGGC---------GGGAAUGAUGGUAACCGCAACAUUUUAGUUGCAUUUGCGACGCUCACCGCAAGAGGCAA------CGCCACAAGGACCAAGGACAUUUACCUAAAU .((((((-.((..((---------((..((....))..)))).))......(((((.((((((........))))))..))))------))))))).......(((.......))).... ( -33.60) >consensus AUGUGGC_AGCAAAU_________GAAAAUGGCGGUAACCGCAACAUUUUAGUUGCAUUUGCAACGUUCGCCACAAGAGGCAA______CGCCACAAGGACCAAGGACAUUUACCUAAAU ..(((((..(((((...................((...))(((((......))))).))))).......(((......))).........)))))........(((.......))).... (-20.41 = -19.93 + -0.47)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:09:29 2006