
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 21,863,777 – 21,863,980 |
| Length | 203 |
| Max. P | 0.980853 |

| Location | 21,863,777 – 21,863,873 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 89.18 |
| Mean single sequence MFE | -39.45 |
| Consensus MFE | -32.62 |
| Energy contribution | -32.50 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.945032 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21863777 96 + 22224390 ACAGUGGGGCAAAACAGUCGGCGGGGGGCGGAUACCGAGACUGUGCUCACGUGAGUACCAGGACAUC--GACUUAGUGUCCUGGGAUUGGUAAUUGCA .((((.((((...((((((..(((..........))).))))))))))((....)).(((((((((.--......))))))))).))))......... ( -36.00) >DroSec_CAF1 55289 96 + 1 ACAGUGGGGCAAAAUAGUCGGCGGGGGGCGGAUGCGGAGACUGUGCUCACGUUAGUGCCAGGACAUC--GACUUAGUGUCCUGGGAUUGGUAAUUGCA ........((((..((((((((((((.((((.........)))).))).))))....(((((((((.--......))))))))))))))....)))). ( -32.70) >DroEre_CAF1 26631 95 + 1 ACAGUGGGGCAAAGCCGUCGGCG-GAGGCGGAUGCCGAGCCUGUGCUCACGUGAGUGCCAGGACACC--GACUUGGUGUCCUGGGAUUGGUAAUUGCA ........((((.((((((((((-.((((.........)))).))))(((....)))((((((((((--.....)))))))))))).))))..)))). ( -44.30) >DroYak_CAF1 28823 98 + 1 ACAGUGAGGCAAAGCAGUCGGCGGGAGGCGGAUACCGAGCCUGUGCUCACGUGAUUGCCAGGACACCAGGUCUUGGUGUCCUGGGACUGGCAAUUGCA ...(((((((...)).....(((((...(((...)))..))))).)))))(..((((((((....(((((.(.....).)))))..))))))))..). ( -44.80) >consensus ACAGUGGGGCAAAACAGUCGGCGGGAGGCGGAUACCGAGACUGUGCUCACGUGAGUGCCAGGACACC__GACUUAGUGUCCUGGGAUUGGUAAUUGCA ........((((..(((((.(((.(((((((.........)))).))).))).....((((((((((.......)))))))))))))))....)))). (-32.62 = -32.50 + -0.12)

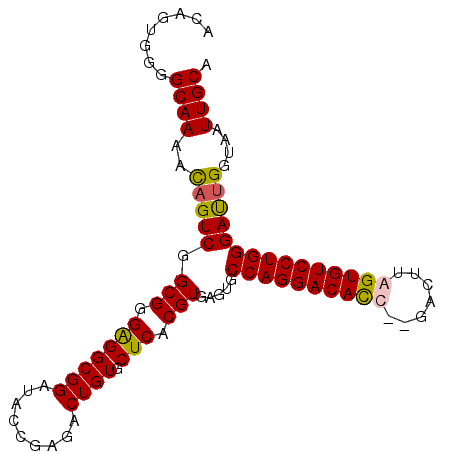

| Location | 21,863,777 – 21,863,873 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 89.18 |
| Mean single sequence MFE | -31.40 |
| Consensus MFE | -25.12 |
| Energy contribution | -25.12 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933632 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21863777 96 - 22224390 UGCAAUUACCAAUCCCAGGACACUAAGUC--GAUGUCCUGGUACUCACGUGAGCACAGUCUCGGUAUCCGCCCCCCGCCGACUGUUUUGCCCCACUGU .((((.........(((((((((......--).))))))))..(((....))).((((((.(((..........)))..)))))).))))........ ( -25.80) >DroSec_CAF1 55289 96 - 1 UGCAAUUACCAAUCCCAGGACACUAAGUC--GAUGUCCUGGCACUAACGUGAGCACAGUCUCCGCAUCCGCCCCCCGCCGACUAUUUUGCCCCACUGU .((...........(((((((((......--).))))))))(((....))).))(((((....(((((.((.....)).))......)))...))))) ( -24.20) >DroEre_CAF1 26631 95 - 1 UGCAAUUACCAAUCCCAGGACACCAAGUC--GGUGUCCUGGCACUCACGUGAGCACAGGCUCGGCAUCCGCCUC-CGCCGACGGCUUUGCCCCACUGU .((...........((((((((((.....--))))))))))(((....))).))((((....((((...(((((-....)).)))..))))...)))) ( -37.20) >DroYak_CAF1 28823 98 - 1 UGCAAUUGCCAGUCCCAGGACACCAAGACCUGGUGUCCUGGCAAUCACGUGAGCACAGGCUCGGUAUCCGCCUCCCGCCGACUGCUUUGCCUCACUGU .((((..((.((((..(((((((((.....)))))))))(((........((((....))))(((....)))....))))))))).))))........ ( -38.40) >consensus UGCAAUUACCAAUCCCAGGACACCAAGUC__GAUGUCCUGGCACUCACGUGAGCACAGGCUCGGCAUCCGCCCCCCGCCGACUGCUUUGCCCCACUGU .(((..........((((((((((.......)))))))))).......(((.(((.((..(((((...........)))))...)).)))..)))))) (-25.12 = -25.12 + 0.00)



| Location | 21,863,835 – 21,863,953 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.20 |
| Mean single sequence MFE | -44.72 |
| Consensus MFE | -33.56 |
| Energy contribution | -34.56 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.06 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928597 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21863835 118 + 22224390 CAGGACAUC--GACUUAGUGUCCUGGGAUUGGUAAUUGCAUUGCCAUGGAUACGCCAACUACGUCUUCGAGUGCAUUACCCGUAGACAACCAGUUGCAUUUGCGUAGGUGGCGCAAUUAA ((((((((.--......)))))))).......(((((((...(((((...((((((((((..((((.((.(((...))).)).))))....))))).....))))).)))))))))))). ( -39.00) >DroSec_CAF1 55347 118 + 1 CAGGACAUC--GACUUAGUGUCCUGGGAUUGGUAAUUGCAUUGCCAUGGAUACGCCAACUGCGUCUUCGAGUGCAUUACUCGUAGACAACCAGUUGCAUUUGCGUAGGUGGCGCAAUUAA ((((((((.--......)))))))).......(((((((...(((((...(((((((((((.((((.((((((...)))))).))))...)))))).....))))).)))))))))))). ( -46.90) >DroEre_CAF1 26688 115 + 1 CAGGACACC--GACUUGGUGUCCUGGGAUUGGUAAUUGCAUUGCCAUGGAUACGC--ACUGCGUCU-CGAGUGCAUUACGCAUAGGCAACCAGUUGCAUUUGCGUAGGUGCCGCAAUUAG (((((((((--.....))))))))).......(((((((..((((........((--(((.((...-)))))))..((((((...((((....))))...))))))))))..))))))). ( -47.80) >DroYak_CAF1 28881 120 + 1 CAGGACACCAGGUCUUGGUGUCCUGGGACUGGCAAUUGCAUUGCCAUGGAUACGACAACUGCGUCUUCAAGUGCAUUACGCGUAGACAACCAGUUGCAUUUGCAUGUGUGGCGCACUUAA (((((((((((...)))))))))))....(((((((...))))))).(((.(((.(....)))).)))((((((.((((((((.(((.....)))((....)))))))))).)))))).. ( -45.20) >consensus CAGGACACC__GACUUAGUGUCCUGGGAUUGGUAAUUGCAUUGCCAUGGAUACGCCAACUGCGUCUUCGAGUGCAUUACGCGUAGACAACCAGUUGCAUUUGCGUAGGUGGCGCAAUUAA (((((((((.......)))))))))....(((((((...)))))))......(((((.(((((....((((((((...................))))))))))))).)))))....... (-33.56 = -34.56 + 1.00)



| Location | 21,863,835 – 21,863,953 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.20 |
| Mean single sequence MFE | -42.30 |
| Consensus MFE | -34.20 |
| Energy contribution | -35.70 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.37 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.980853 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21863835 118 - 22224390 UUAAUUGCGCCACCUACGCAAAUGCAACUGGUUGUCUACGGGUAAUGCACUCGAAGACGUAGUUGGCGUAUCCAUGGCAAUGCAAUUACCAAUCCCAGGACACUAAGUC--GAUGUCCUG .(((((((((((..(((((.....((((((..(((((.(((((.....))))).))))))))))))))))....))))...))))))).......((((((((......--).))))))) ( -39.70) >DroSec_CAF1 55347 118 - 1 UUAAUUGCGCCACCUACGCAAAUGCAACUGGUUGUCUACGAGUAAUGCACUCGAAGACGCAGUUGGCGUAUCCAUGGCAAUGCAAUUACCAAUCCCAGGACACUAAGUC--GAUGUCCUG .(((((((((((..(((((.....((((((..(((((.(((((.....))))).))))))))))))))))....))))...))))))).......((((((((......--).))))))) ( -42.20) >DroEre_CAF1 26688 115 - 1 CUAAUUGCGGCACCUACGCAAAUGCAACUGGUUGCCUAUGCGUAAUGCACUCG-AGACGCAGU--GCGUAUCCAUGGCAAUGCAAUUACCAAUCCCAGGACACCAAGUC--GGUGUCCUG .((((((((...((((((((...((((....))))...))))))(((((((((-...)).)))--))))......))...)))))))).......(((((((((.....--))))))))) ( -45.20) >DroYak_CAF1 28881 120 - 1 UUAAGUGCGCCACACAUGCAAAUGCAACUGGUUGUCUACGCGUAAUGCACUUGAAGACGCAGUUGUCGUAUCCAUGGCAAUGCAAUUGCCAGUCCCAGGACACCAAGACCUGGUGUCCUG ....(((.....))).(((....)))((((((.((((.((.((.....)).)).))))(((.(((((((....))))))))))....))))))..((((((((((.....)))))))))) ( -42.10) >consensus UUAAUUGCGCCACCUACGCAAAUGCAACUGGUUGUCUACGCGUAAUGCACUCGAAGACGCAGUUGGCGUAUCCAUGGCAAUGCAAUUACCAAUCCCAGGACACCAAGUC__GAUGUCCUG .(((((((((((..(((((.....((((((..(((((.(((((.....))))).))))))))))))))))....))))...))))))).......(((((((((.......))))))))) (-34.20 = -35.70 + 1.50)


| Location | 21,863,873 – 21,863,980 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 88.16 |
| Mean single sequence MFE | -33.64 |
| Consensus MFE | -21.24 |
| Energy contribution | -22.05 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.542250 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21863873 107 - 22224390 CCAGCUUAGUGCUGUAAGCAACACUUCUUAAUUGCGCCACCUACGCAAAUGCAACUGGUUGUCUACGGGUAAUGCACUCGAAGACGUAGUUGGCGUAUCCAUGGCAA ...(((.((((.((....)).)))).......(((((((.((((((....))........((((.(((((.....))))).)))))))).))))))).....))).. ( -35.10) >DroSec_CAF1 55385 107 - 1 CAAGCUUAGUGCCGUAAGCAACACUUGUUAAUUGCGCCACCUACGCAAAUGCAACUGGUUGUCUACGAGUAAUGCACUCGAAGACGCAGUUGGCGUAUCCAUGGCAA .........((((((.((((.....))))...((((((......((....))(((((..(((((.(((((.....))))).))))))))))))))))...)))))). ( -35.30) >DroEre_CAF1 26726 104 - 1 CCAGCUUCAUGCCGCAAGCAACACUUCCUAAUUGCGGCACCUACGCAAAUGCAACUGGUUGCCUAUGCGUAAUGCACUCG-AGACGCAGU--GCGUAUCCAUGGCAA ((((.....((((((((..............)))))))).....((....))..))))(((((.(((.(((.((((((((-...)).)))--)))))).)))))))) ( -34.74) >DroYak_CAF1 28921 107 - 1 CCAGCUUCAUGCGGCUAGCAACUCUUCUUAAGUGCGCCACACAUGCAAAUGCAACUGGUUGUCUACGCGUAAUGCACUUGAAGACGCAGUUGUCGUAUCCAUGGCAA ...(((..(((((((.(((..((((((..((((((((((....(((....)))..)))).((....)).....))))))))))).)..))))))))))....))).. ( -29.40) >consensus CCAGCUUAAUGCCGCAAGCAACACUUCUUAAUUGCGCCACCUACGCAAAUGCAACUGGUUGUCUACGCGUAAUGCACUCGAAGACGCAGUUGGCGUAUCCAUGGCAA .........(((((.................(((((.......)))))(((((((((..(((((.(((((.....))))).))))))))))).))).....))))). (-21.24 = -22.05 + 0.81)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:09:26 2006