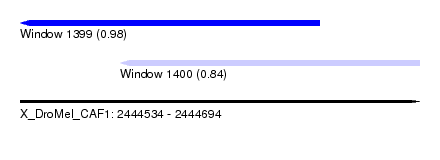
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,444,534 – 2,444,694 |
| Length | 160 |
| Max. P | 0.977331 |

| Location | 2,444,534 – 2,444,654 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.54 |
| Mean single sequence MFE | -29.16 |
| Consensus MFE | -27.78 |
| Energy contribution | -27.34 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977331 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
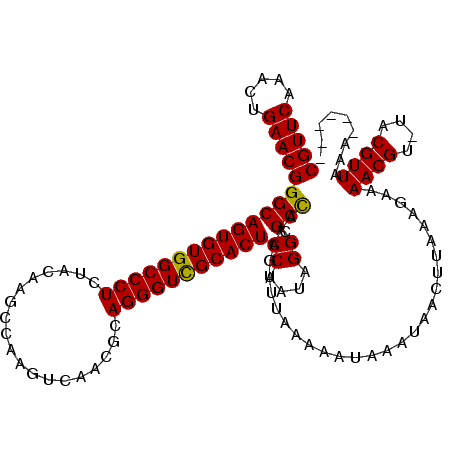
>X_DroMel_CAF1 2444534 120 - 22224390 CGUUCAAACUGAACGGGCAGUGUGGCCCUCUUCAACCGAACUCAACGCAGGGUCGCACUCCAAUAGGCGCUAGAGUUUAAAAAUAAAUAACUUAAAGAAAACGUCUACGUUAAGUAUUAU .......(((.((((((.((((((((((((................).)))))))))))))..((((((((.(((((...........)))))..))....)))))))))).)))..... ( -30.79) >DroSec_CAF1 5375 112 - 1 CGUUCAAACUGAACGGGCAGUGUGGCCCUCUAUAAGCCAAGUCAACGCAGGGUCGCACUCCAAUAGGCGCCAAAAC-UAAAAAUAAAUAACUUAAAGAAAACGU-UACGUUAAA------ (((((.....)))))((((((((((((((......((.........)))))))))))))((....)).))).....-..........((((.(((........)-)).))))..------ ( -27.60) >DroSim_CAF1 7557 113 - 1 CGUUCAAACUGAACGGGCAGUGUGGCCCUCUACAAGGCAAGUCAGCUCAGGGUUGCACUCCAAUAGGCGCCAAAGUUUAAAAAUAAAUAACUUAAAGAAAACGU-UACGUUAAA------ (((((.....)))))((.((((..(((((......(((......))).)))))..))))))..(((((......)))))........((((.(((........)-)).))))..------ ( -29.10) >consensus CGUUCAAACUGAACGGGCAGUGUGGCCCUCUACAAGCCAAGUCAACGCAGGGUCGCACUCCAAUAGGCGCCAAAGUUUAAAAAUAAAUAACUUAAAGAAAACGU_UACGUUAAA______ (((((.....)))))((((((((((((((...................)))))))))))((....)).)))............................((((....))))......... (-27.78 = -27.34 + -0.44)

| Location | 2,444,574 – 2,444,694 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.11 |
| Mean single sequence MFE | -40.83 |
| Consensus MFE | -38.06 |
| Energy contribution | -37.84 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.837268 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
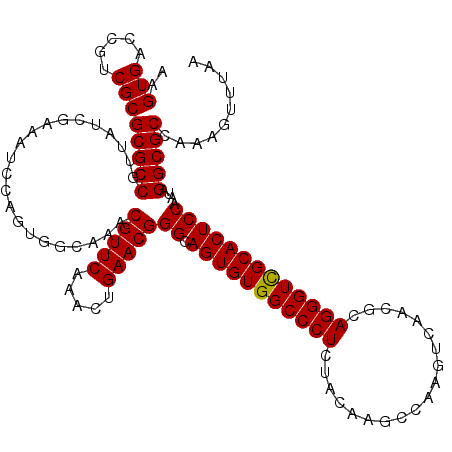
>X_DroMel_CAF1 2444574 120 - 22224390 AAGUGACCGUCGCGCGCCGUUGUCGAAAUCCAGUGGCAAACGUUCAAACUGAACGGGCAGUGUGGCCCUCUUCAACCGAACUCAACGCAGGGUCGCACUCCAAUAGGCGCUAGAGUUUAA ..(((.....)))(((((.((((((........)))))).(((((.....)))))((.((((((((((((................).)))))))))))))....))))).......... ( -42.29) >DroSec_CAF1 5408 119 - 1 AAGUGACCGUCGCGCGCCGUUAUCGAAAUCCAGUGGCAAACGUUCAAACUGAACGGGCAGUGUGGCCCUCUAUAAGCCAAGUCAACGCAGGGUCGCACUCCAAUAGGCGCCAAAAC-UAA ..(((.....)))((((((((((.(.....).)))))...(((((.....)))))((.(((((((((((......((.........)))))))))))))))....)))))......-... ( -40.50) >DroSim_CAF1 7590 120 - 1 AAGUGACCGUCGCGCGCCGUUAUCGAAAUCCAGUGCCAAACGUUCAAACUGAACGGGCAGUGUGGCCCUCUACAAGGCAAGUCAGCUCAGGGUUGCACUCCAAUAGGCGCCAAAGUUUAA ..(((.....)))(((((......................(((((.....)))))((.((((..(((((......(((......))).)))))..))))))....))))).......... ( -39.70) >consensus AAGUGACCGUCGCGCGCCGUUAUCGAAAUCCAGUGGCAAACGUUCAAACUGAACGGGCAGUGUGGCCCUCUACAAGCCAAGUCAACGCAGGGUCGCACUCCAAUAGGCGCCAAAGUUUAA ..(((.....)))(((((......................(((((.....)))))((.(((((((((((...................)))))))))))))....))))).......... (-38.06 = -37.84 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:55:42 2006