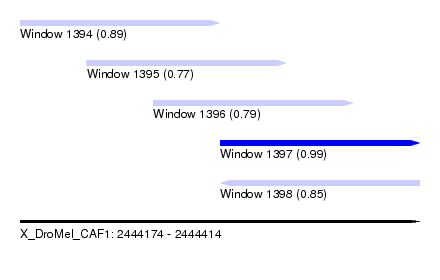
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,444,174 – 2,444,414 |
| Length | 240 |
| Max. P | 0.993847 |

| Location | 2,444,174 – 2,444,294 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -25.30 |
| Consensus MFE | -22.07 |
| Energy contribution | -22.40 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.893360 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2444174 120 + 22224390 AACAUAACUUAUAAUGAUCUUUCGGUGACGAGAAAACUAAAUUUAAGCGUAAAAGAUACAACAGGUGCUCGGGGCAGAAUCCCAGCGCCAGAAAGAAUAUAUAUAAAAUACGAAAUGUAA ........((((((((.(((((((....)..................................((((((.((((.....)))))))))).)))))).))).))))).............. ( -25.80) >DroSec_CAF1 5036 120 + 1 AACAAAACUUAUAAUGAUCUUUCGGUGACGAGAAAACUAAAUUUAAGCGAAACACAUACAACAGGUGCUCGGGGCAGAAUCCCAGCGCCAGAAAGAAUAUAUAUAAAAUACGAAAUGUAA ........((((((((.(((((((....)..................................((((((.((((.....)))))))))).)))))).))).))))).............. ( -25.80) >DroSim_CAF1 7219 120 + 1 AACAAAACUUACAAUGAUCUUUCGGUGACGAGAAAACUAAAUUUAAGUGAAACACAUACAACAGGUGCUCGGGUCAGAAUCCCAGCGCCAGAAAGAAUAUAUAUAAAAUACGAAAUGUAA .........((((......(((((....))))).......((((..(((...)))........((((((.(((.......))))))))).....)))).................)))). ( -24.30) >consensus AACAAAACUUAUAAUGAUCUUUCGGUGACGAGAAAACUAAAUUUAAGCGAAACACAUACAACAGGUGCUCGGGGCAGAAUCCCAGCGCCAGAAAGAAUAUAUAUAAAAUACGAAAUGUAA ........((((((((.(((((((((.........))).........................((((((.(((.......))))))))).)))))).))).))))).............. (-22.07 = -22.40 + 0.33)

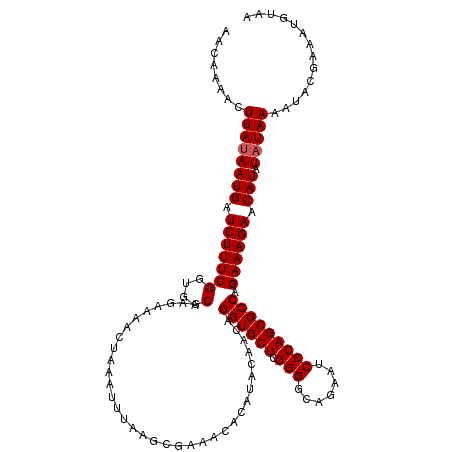
| Location | 2,444,214 – 2,444,334 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.56 |
| Mean single sequence MFE | -25.30 |
| Consensus MFE | -23.59 |
| Energy contribution | -23.37 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.772894 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2444214 120 + 22224390 AUUUAAGCGUAAAAGAUACAACAGGUGCUCGGGGCAGAAUCCCAGCGCCAGAAAGAAUAUAUAUAAAAUACGAAAUGUAAAUUUUGAAUAAAAGAAAACACCGUCUCGGCGGCGCCCAAG ......((((....((.((....((((((.((((.....)))))))))).....................((((((....))))))................)).))....))))..... ( -27.30) >DroSec_CAF1 5076 120 + 1 AUUUAAGCGAAACACAUACAACAGGUGCUCGGGGCAGAAUCCCAGCGCCAGAAAGAAUAUAUAUAAAAUACGAAAUGUAAAUUUUGAAUAAAAGAAAACACCGUCUCAGCGGCGCCCAAA ......(((..............((((((.((((.....)))))))))).....................((((((....))))))..............((((....)))))))..... ( -25.30) >DroSim_CAF1 7259 120 + 1 AUUUAAGUGAAACACAUACAACAGGUGCUCGGGUCAGAAUCCCAGCGCCAGAAAGAAUAUAUAUAAAAUACGAAAUGUAAAUUUUGAAUAAAAGAAAACACCGUCUCAUCGGCGCCCAAG ......(((..............((((((.(((.......))))))))).....................((((((....))))))............)))((((.....))))...... ( -23.30) >consensus AUUUAAGCGAAACACAUACAACAGGUGCUCGGGGCAGAAUCCCAGCGCCAGAAAGAAUAUAUAUAAAAUACGAAAUGUAAAUUUUGAAUAAAAGAAAACACCGUCUCAGCGGCGCCCAAG ......(((..............((((((.(((.......))))))))).....................((((((....))))))..............(((......))))))..... (-23.59 = -23.37 + -0.22)

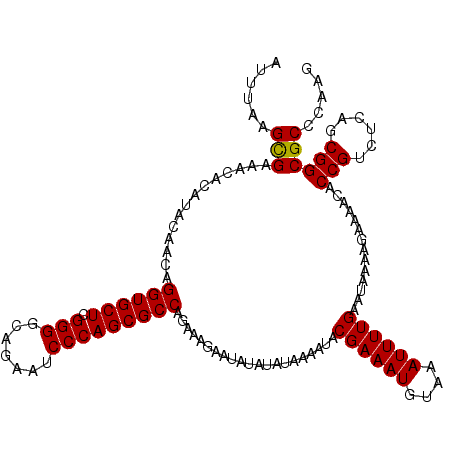
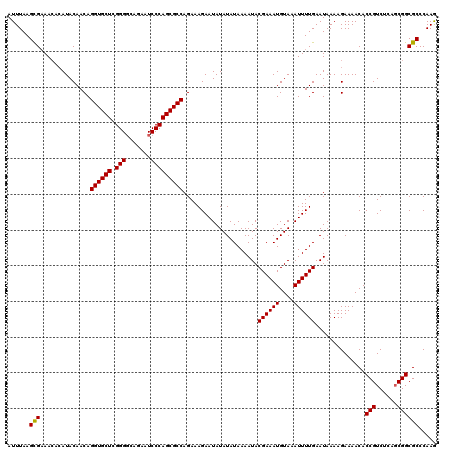
| Location | 2,444,254 – 2,444,374 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -28.00 |
| Consensus MFE | -28.03 |
| Energy contribution | -27.70 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.67 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.790852 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2444254 120 + 22224390 CCCAGCGCCAGAAAGAAUAUAUAUAAAAUACGAAAUGUAAAUUUUGAAUAAAAGAAAACACCGUCUCGGCGGCGCCCAAGUGUCCUUUCAACUUGCGCGCUUUUCGAGGCGAGACACUUA ............(((...............((((((....))))))...............((((((((.(((((.(((((.........))))).)))))..)))))))).....))). ( -29.10) >DroSec_CAF1 5116 120 + 1 CCCAGCGCCAGAAAGAAUAUAUAUAAAAUACGAAAUGUAAAUUUUGAAUAAAAGAAAACACCGUCUCAGCGGCGCCCAAAUGUCCUUUCAACUUGCGCGCCUUUUGAGGCGAGACACUUA ............(((...............((((((....))))))...............((((((((.(((((.(((.((......))..))).)))))..)))))))).....))). ( -25.00) >DroSim_CAF1 7299 120 + 1 CCCAGCGCCAGAAAGAAUAUAUAUAAAAUACGAAAUGUAAAUUUUGAAUAAAAGAAAACACCGUCUCAUCGGCGCCCAAGUGUCCUUUCAACUUGCGCGCCUUUUGAGGCGAGACACUUA ............(((...............((((((....))))))...............(((((((..(((((.(((((.........))))).)))))...))))))).....))). ( -29.90) >consensus CCCAGCGCCAGAAAGAAUAUAUAUAAAAUACGAAAUGUAAAUUUUGAAUAAAAGAAAACACCGUCUCAGCGGCGCCCAAGUGUCCUUUCAACUUGCGCGCCUUUUGAGGCGAGACACUUA ............(((...............((((((....))))))...............(((((((..(((((.(((((.........))))).)))))...))))))).....))). (-28.03 = -27.70 + -0.33)

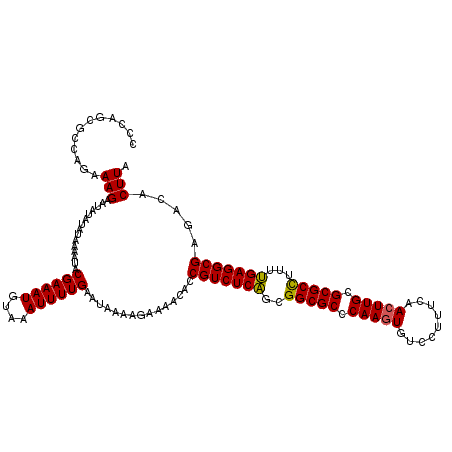
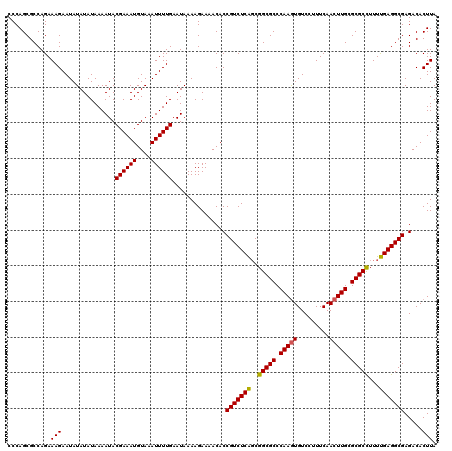
| Location | 2,444,294 – 2,444,414 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.85 |
| Mean single sequence MFE | -31.93 |
| Consensus MFE | -31.83 |
| Energy contribution | -31.50 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.82 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.43 |
| SVM RNA-class probability | 0.993847 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2444294 120 + 22224390 AUUUUGAAUAAAAGAAAACACCGUCUCGGCGGCGCCCAAGUGUCCUUUCAACUUGCGCGCUUUUCGAGGCGAGACACUUAAAUGUACAAAUUGCAUUUUAUUCACAUAAUAAGAGAAGUU ....(((((((((........((((((((.(((((.(((((.........))))).)))))..))))))))...........((((.....)))))))))))))................ ( -32.90) >DroSec_CAF1 5156 118 + 1 AUUUUGAAUAAAAGAAAACACCGUCUCAGCGGCGCCCAAAUGUCCUUUCAACUUGCGCGCCUUUUGAGGCGAGACACUUAAAUGUACAAAUUGCAUUUUAUUCACUCAAUG--AGAAGUU ....(((((((((........((((((((.(((((.(((.((......))..))).)))))..))))))))...........((((.....)))))))))))))(((...)--))..... ( -29.20) >DroSim_CAF1 7339 118 + 1 AUUUUGAAUAAAAGAAAACACCGUCUCAUCGGCGCCCAAGUGUCCUUUCAACUUGCGCGCCUUUUGAGGCGAGACACUUAAAUGUACAAAUUGCAUUUUAUUCACUCGAUA--AGAAGUU ....(((((((((........(((((((..(((((.(((((.........))))).)))))...)))))))...........((((.....))))))))))))).......--....... ( -33.70) >consensus AUUUUGAAUAAAAGAAAACACCGUCUCAGCGGCGCCCAAGUGUCCUUUCAACUUGCGCGCCUUUUGAGGCGAGACACUUAAAUGUACAAAUUGCAUUUUAUUCACUCAAUA__AGAAGUU ....(((((((((........(((((((..(((((.(((((.........))))).)))))...)))))))...........((((.....)))))))))))))................ (-31.83 = -31.50 + -0.33)

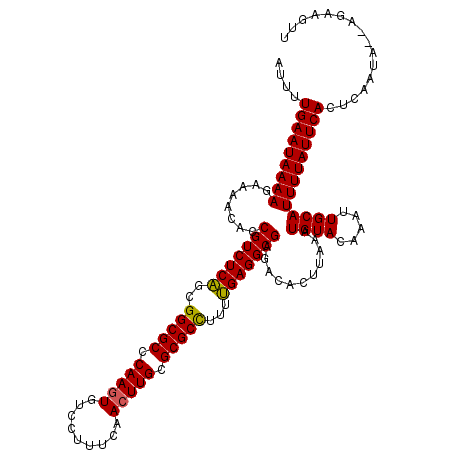
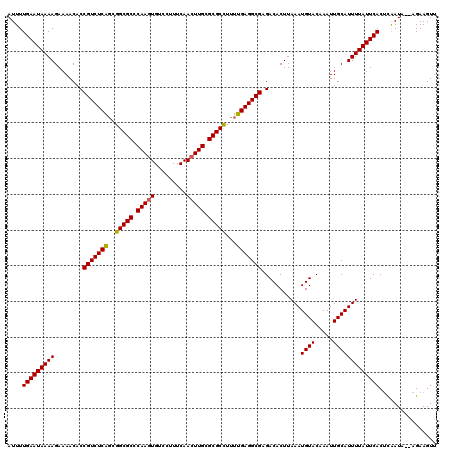
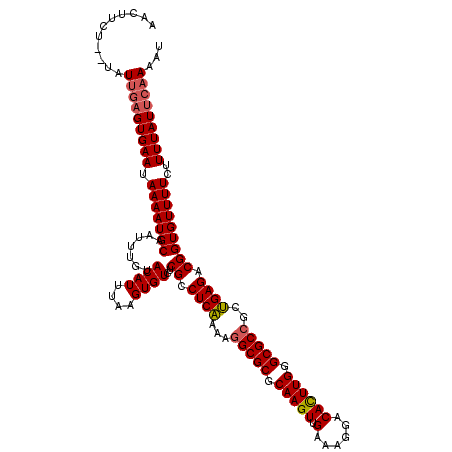
| Location | 2,444,294 – 2,444,414 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.85 |
| Mean single sequence MFE | -32.33 |
| Consensus MFE | -30.70 |
| Energy contribution | -31.37 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.846072 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2444294 120 - 22224390 AACUUCUCUUAUUAUGUGAAUAAAAUGCAAUUUGUACAUUUAAGUGUCUCGCCUCGAAAAGCGCGCAAGUUGAAAGGACACUUGGGCGCCGCCGAGACGGUGUUUUCUUUUAUUCAAAAU ................(((((((((.((((((((......)))))((((((...((....((((.(((((.(......)))))).)))))).))))))..)))....))))))))).... ( -29.10) >DroSec_CAF1 5156 118 - 1 AACUUCU--CAUUGAGUGAAUAAAAUGCAAUUUGUACAUUUAAGUGUCUCGCCUCAAAAGGCGCGCAAGUUGAAAGGACAUUUGGGCGCCGCUGAGACGGUGUUUUCUUUUAUUCAAAAU .......--..(((((((((.(((((((.......((((....))))..((.((((...(((((.(((((.(......)))))).)))))..)))).)))))))))..)))))))))... ( -33.60) >DroSim_CAF1 7339 118 - 1 AACUUCU--UAUCGAGUGAAUAAAAUGCAAUUUGUACAUUUAAGUGUCUCGCCUCAAAAGGCGCGCAAGUUGAAAGGACACUUGGGCGCCGAUGAGACGGUGUUUUCUUUUAUUCAAAAU .......--....(((((((.(((((((.......((((....))))..((.((((...(((((.(((((.(......)))))).)))))..)))).)))))))))..)))))))..... ( -34.30) >consensus AACUUCU__UAUUGAGUGAAUAAAAUGCAAUUUGUACAUUUAAGUGUCUCGCCUCAAAAGGCGCGCAAGUUGAAAGGACACUUGGGCGCCGCUGAGACGGUGUUUUCUUUUAUUCAAAAU ...........(((((((((.(((((((.......((((....))))..((.((((...(((((.(((((.(......)))))).)))))..)))).)))))))))..)))))))))... (-30.70 = -31.37 + 0.67)

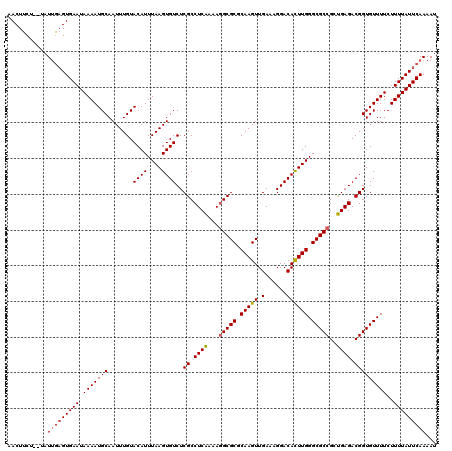
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:55:40 2006