
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 21,702,300 – 21,702,498 |
| Length | 198 |
| Max. P | 0.995074 |

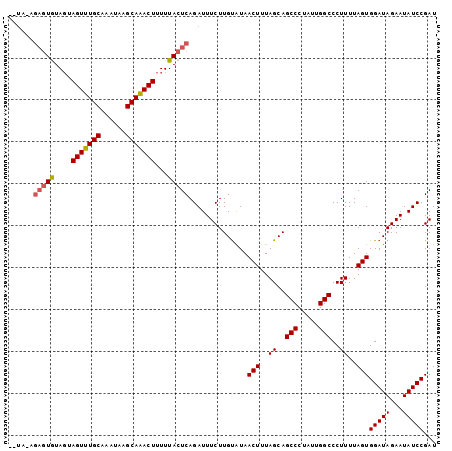
| Location | 21,702,300 – 21,702,402 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 88.49 |
| Mean single sequence MFE | -24.10 |
| Consensus MFE | -23.66 |
| Energy contribution | -23.00 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.54 |
| SVM RNA-class probability | 0.995074 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21702300 102 + 22224390 AUCGGAUAUUCUAUCCACUAAAAGGGCCAAGAAGGCUGCUAAAGUUACGCAAGAAAUCUGAGUAAAAAGUGUGCUUAUUUGCACACUACUAUACUCUCUAUA ...(((((...)))))........((((.....))))..........(....)......(((((...(((((((......)))))))....)))))...... ( -26.20) >DroSec_CAF1 6946 100 + 1 AUCGGAUAUUCUAUCCACUAAAAGGGCCAAUAGGGCUGCUAAAGUUAUACAAGAAAUCUGAGUAAAAAGUUUGCUUAUUUGCAAACUACUACACUCUGUA-- .((((((.((((....(((...((((((.....)))).))..)))......))))))))))(((...(((((((......)))))))..)))........-- ( -22.50) >DroSim_CAF1 3749 100 + 1 AUCGGAUAUUCUAUCCACUAAAAGGGCCAAUAGGGCUGCUAAAGUUAUACAAGAAAUCUGAGUAAAAAGUUUGCUUAUUUGCAAACUAGUACANNNNNNN-- .((((((.((((....(((...((((((.....)))).))..)))......))))))))))(((...(((((((......)))))))..)))........-- ( -23.60) >consensus AUCGGAUAUUCUAUCCACUAAAAGGGCCAAUAGGGCUGCUAAAGUUAUACAAGAAAUCUGAGUAAAAAGUUUGCUUAUUUGCAAACUACUACACUCU_UA__ .((((((.((((....(((...((((((.....)))).))..)))......))))))))))(((...(((((((......)))))))..))).......... (-23.66 = -23.00 + -0.66)



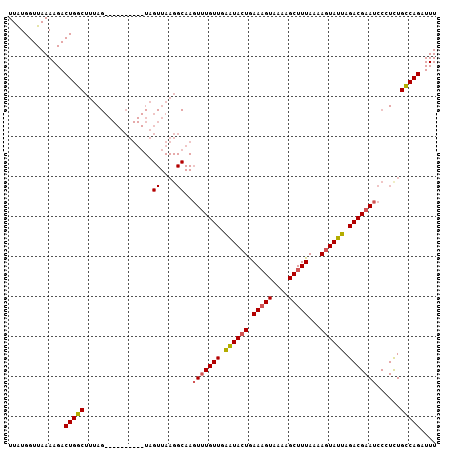
| Location | 21,702,300 – 21,702,402 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 88.49 |
| Mean single sequence MFE | -22.93 |
| Consensus MFE | -20.76 |
| Energy contribution | -21.10 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.763727 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21702300 102 - 22224390 UAUAGAGAGUAUAGUAGUGUGCAAAUAAGCACACUUUUUACUCAGAUUUCUUGCGUAACUUUAGCAGCCUUCUUGGCCCUUUUAGUGGAUAGAAUAUCCGAU ......(((((....(((((((......)))))))...)))))........(((.........)))(((.....)))........((((((...)))))).. ( -27.10) >DroSec_CAF1 6946 100 - 1 --UACAGAGUGUAGUAGUUUGCAAAUAAGCAAACUUUUUACUCAGAUUUCUUGUAUAACUUUAGCAGCCCUAUUGGCCCUUUUAGUGGAUAGAAUAUCCGAU --(((((((((....(((((((......)))))))...)))))((....))))))..(((..((..(((.....)))..))..)))(((((...)))))... ( -22.90) >DroSim_CAF1 3749 100 - 1 --NNNNNNNUGUACUAGUUUGCAAAUAAGCAAACUUUUUACUCAGAUUUCUUGUAUAACUUUAGCAGCCCUAUUGGCCCUUUUAGUGGAUAGAAUAUCCGAU --........(((..(((((((......)))))))...)))((.(((((((.((...(((..((..(((.....)))..))..)))..)))))).))).)). ( -18.80) >consensus __UA_AGAGUGUAGUAGUUUGCAAAUAAGCAAACUUUUUACUCAGAUUUCUUGUAUAACUUUAGCAGCCCUAUUGGCCCUUUUAGUGGAUAGAAUAUCCGAU ......(((((....(((((((......)))))))...)))))..............(((..((..(((.....)))..))..)))(((((...)))))... (-20.76 = -21.10 + 0.34)


| Location | 21,702,402 – 21,702,498 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 75.56 |
| Mean single sequence MFE | -20.20 |
| Consensus MFE | -14.19 |
| Energy contribution | -15.03 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.558067 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21702402 96 + 22224390 UUGUGGUUAAAAGACUGGCUUUAG----------UAGUUAAGGCAAGU-UGUUGAAUACUGAAAGUAAAAGCUUUAAAAGUAUUAGACGAAUCCGUUUGUCAGAUUU ............((((.(((((((----------...))))))).)))-)....((((((.(((((....)))))...)))))).((((((....))))))...... ( -20.40) >DroSec_CAF1 7046 107 + 1 UUAUGGUUAAAAGACUGGCUUUAGUGAUUCUUUGUAGUUAAGGCAAGUUUGUUGGAUACUAAAAGUAAAAGCUUUAAAAGUAUUAGACGAAUCCCUCUGCCAGAUUU ..............(((((...((.((((((((((.......)))))...(((.((((((.(((((....)))))...)))))).)))))))).))..))))).... ( -25.20) >DroEre_CAF1 4639 83 + 1 UUAUGGUUAAAAGACUGGCCUUA-----------------------CUUUGUUAAGUACUGAA-GUAAUAGCUUUAAAAAUAUUAGACGAAUCCCUCUGCCAGGUUU ..............(((((....-----------------------.((((((.((((.((((-((....))))))....)))).)))))).......))))).... ( -15.00) >consensus UUAUGGUUAAAAGACUGGCUUUAG__________UAGUUAAGGCAAGUUUGUUGAAUACUGAAAGUAAAAGCUUUAAAAGUAUUAGACGAAUCCCUCUGCCAGAUUU ..............(((((.................((....))..(((((((.((((((.(((((....)))))...)))))).)))))))......))))).... (-14.19 = -15.03 + 0.84)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:08:58 2006