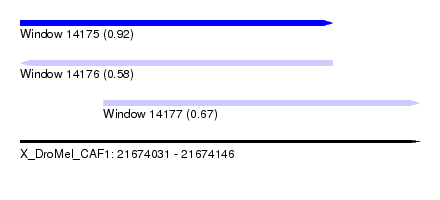
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 21,674,031 – 21,674,146 |
| Length | 115 |
| Max. P | 0.922048 |

| Location | 21,674,031 – 21,674,121 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 81.85 |
| Mean single sequence MFE | -34.72 |
| Consensus MFE | -26.98 |
| Energy contribution | -30.15 |
| Covariance contribution | 3.17 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922048 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
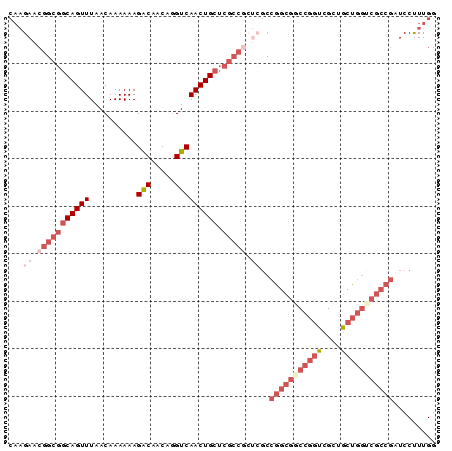
>X_DroMel_CAF1 21674031 90 + 22224390 CAAGAACGGCGGCAGUUUAACAAAAAAGACAACAGGUCAACUGCUCGCCGCUCGCCGGCGUCCGGUCGCUGCUGGUCGCCGAUCCCUUGG ((((..(((((((((((..........(((.....))))))))).))))).....(((((.(((((....))))).)))))....)))). ( -38.10) >DroSec_CAF1 4433 83 + 1 CAAGAACGGCGGCAGUUUAACAAAAAAGACAACAGGUCAACUGCUC-------GCCGGCGGCCGGUCGCUGCUGGCCGCCGAUCCUUUGG ((((...((((((((((..........(((.....))))))))).)-------)))((((((((((....)))))))))).....)))). ( -38.50) >DroSim_CAF1 5001 90 + 1 CAAGAACGGCGGCAGUUUAACAAAAAAGACAACAGGUCAACUGCUCGCCGCUCGCCGGCGGCCGGUCGCUGCUGGUCGCCGAUCCUUUGG (((((.(((((((((((..........(((.....))))))))).))))).))..(((((((((((....))))))))))).....))). ( -40.70) >DroEre_CAF1 4432 90 + 1 CAAGAACGGCGGCAGUUUAACAAAAAAGACAACAGGUCAACUGCUCGCCGCUCGCCGGCGGCCGGUCGCUGCUGGUCGCCGAUCCUUUGG (((((.(((((((((((..........(((.....))))))))).))))).))..(((((((((((....))))))))))).....))). ( -40.70) >DroYak_CAF1 5198 90 + 1 CAAGAACGGCGGCAGUUUAACAAAAAAGACAACAGGUCAACUGCUCGCCGCUCGCCGGCGGCCGGUCGCUGCUGGUCGCCGAUCCUUUGG (((((.(((((((((((..........(((.....))))))))).))))).))..(((((((((((....))))))))))).....))). ( -40.70) >DroPer_CAF1 5354 71 + 1 CAA--AAGGCAACAGUUUAACAAAAAAGGCAACCAGUCAACUGAGCGCCACUUGCAUGCACCGAAACGUUUCG----------------- ...--...((((..(.....)......(((...(((....)))...)))..))))......((((....))))----------------- ( -9.60) >consensus CAAGAACGGCGGCAGUUUAACAAAAAAGACAACAGGUCAACUGCUCGCCGCUCGCCGGCGGCCGGUCGCUGCUGGUCGCCGAUCCUUUGG ...((.(((((((((((..........(((.....))))))))).))))).))..(((((((((((....)))))))))))......... (-26.98 = -30.15 + 3.17)


| Location | 21,674,031 – 21,674,121 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 81.85 |
| Mean single sequence MFE | -33.02 |
| Consensus MFE | -22.89 |
| Energy contribution | -25.78 |
| Covariance contribution | 2.89 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.576392 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
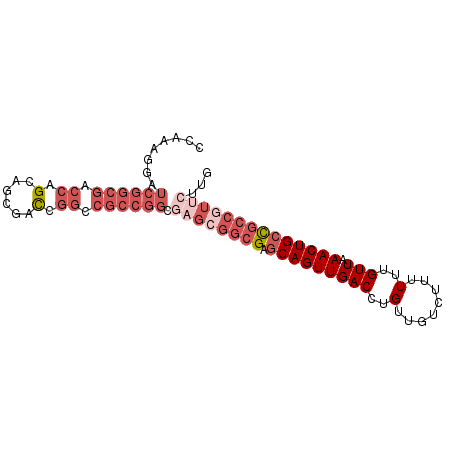
>X_DroMel_CAF1 21674031 90 - 22224390 CCAAGGGAUCGGCGACCAGCAGCGACCGGACGCCGGCGAGCGGCGAGCAGUUGACCUGUUGUCUUUUUUGUUAAACUGCCGCCGUUCUUG ......(.((((((.((.(......).)).)))))))((((((((.(((((((((..(........)..))).))))))))))))))... ( -34.90) >DroSec_CAF1 4433 83 - 1 CCAAAGGAUCGGCGGCCAGCAGCGACCGGCCGCCGGC-------GAGCAGUUGACCUGUUGUCUUUUUUGUUAAACUGCCGCCGUUCUUG ...((((((.(((((((.(......).)))))))(((-------(.(((((((((..(........)..))).)))))))))))))))). ( -35.70) >DroSim_CAF1 5001 90 - 1 CCAAAGGAUCGGCGACCAGCAGCGACCGGCCGCCGGCGAGCGGCGAGCAGUUGACCUGUUGUCUUUUUUGUUAAACUGCCGCCGUUCUUG ......(.((((((.((.(......).)).)))))))((((((((.(((((((((..(........)..))).))))))))))))))... ( -35.30) >DroEre_CAF1 4432 90 - 1 CCAAAGGAUCGGCGACCAGCAGCGACCGGCCGCCGGCGAGCGGCGAGCAGUUGACCUGUUGUCUUUUUUGUUAAACUGCCGCCGUUCUUG ......(.((((((.((.(......).)).)))))))((((((((.(((((((((..(........)..))).))))))))))))))... ( -35.30) >DroYak_CAF1 5198 90 - 1 CCAAAGGAUCGGCGACCAGCAGCGACCGGCCGCCGGCGAGCGGCGAGCAGUUGACCUGUUGUCUUUUUUGUUAAACUGCCGCCGUUCUUG ......(.((((((.((.(......).)).)))))))((((((((.(((((((((..(........)..))).))))))))))))))... ( -35.30) >DroPer_CAF1 5354 71 - 1 -----------------CGAAACGUUUCGGUGCAUGCAAGUGGCGCUCAGUUGACUGGUUGCCUUUUUUGUUAAACUGUUGCCUU--UUG -----------------(((((.((..((((....(((((.((((..(((....)))..))))...)))))...))))..)).))--))) ( -21.60) >consensus CCAAAGGAUCGGCGACCAGCAGCGACCGGCCGCCGGCGAGCGGCGAGCAGUUGACCUGUUGUCUUUUUUGUUAAACUGCCGCCGUUCUUG ........((((((.((.(......).)).)))))).((((((((.(((((((((..(........)..))).))))))))))))))... (-22.89 = -25.78 + 2.89)


| Location | 21,674,055 – 21,674,146 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 93.74 |
| Mean single sequence MFE | -40.16 |
| Consensus MFE | -35.70 |
| Energy contribution | -36.54 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.672459 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
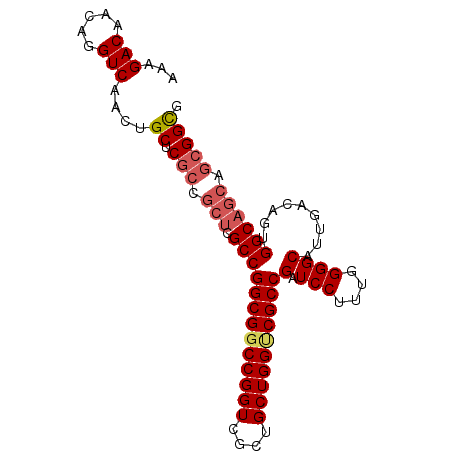
>X_DroMel_CAF1 21674055 91 + 22224390 AAAGACAACAGGUCAACUGCUCGCCGCUCGCCGGCGUCCGGUCGCUGCUGGUCGCCGAUCCCUUGGGGCAUUGACAGUGGCAGCAGCGGCG ...........(((..(((((.((((((...(((((.(((((....))))).))))).(((....))).......))))))))))).))). ( -39.10) >DroSec_CAF1 4457 84 + 1 AAAGACAACAGGUCAACUGCUC-------GCCGGCGGCCGGUCGCUGCUGGCCGCCGAUCCUUUGGGGCAUUGACAGUGGCAGCAGCGGCG ...........(((..(((((.-------(((((((((((((....))))))))))(.(((....)))).........)))))))).))). ( -38.40) >DroSim_CAF1 5025 91 + 1 AAAGACAACAGGUCAACUGCUCGCCGCUCGCCGGCGGCCGGUCGCUGCUGGUCGCCGAUCCUUUGGGGCAUUGACAGUGGCAGCAGCGGCG ...........(((..(((((.((((((...(((((((((((....))))))))))).(((....))).......))))))))))).))). ( -41.30) >DroEre_CAF1 4456 91 + 1 AAAGACAACAGGUCAACUGCUCGCCGCUCGCCGGCGGCCGGUCGCUGCUGGUCGCCGAUCCUUUGGGGCAUCGACAAUGGCAGCAGCGGUG ...(((.....))).......(((((((((((((((((((((....))))))))))(((((.....)).)))......))).).))))))) ( -40.90) >DroYak_CAF1 5222 91 + 1 AAAGACAACAGGUCAACUGCUCGCCGCUCGCCGGCGGCCGGUCGCUGCUGGUCGCCGAUCCUUUGGGGCAUAGACAGUGGCAGCAGCGGUG ...(((.....)))..(((((.((((((...(((((((((((....))))))))))).(((....))).......)))))))))))..... ( -41.10) >consensus AAAGACAACAGGUCAACUGCUCGCCGCUCGCCGGCGGCCGGUCGCUGCUGGUCGCCGAUCCUUUGGGGCAUUGACAGUGGCAGCAGCGGCG ...(((.....)))....((.(((.(((.(((((((((((((....))))))))))(.(((....)))).........)))))).))))). (-35.70 = -36.54 + 0.84)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:08:48 2006