
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 21,648,736 – 21,648,836 |
| Length | 100 |
| Max. P | 0.984251 |

| Location | 21,648,736 – 21,648,836 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 78.54 |
| Mean single sequence MFE | -31.65 |
| Consensus MFE | -26.25 |
| Energy contribution | -25.62 |
| Covariance contribution | -0.62 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.984251 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21648736 100 + 22224390 AUCCCAACACGUCAGCAGA-----AUCUGUGUGAUGUGCUUGGGAGGAAAUUUCCAAGCGGCA-AGAUGUGAUAACGAUCGUGUCGCUGAUAUGUA----CUUGGC-----AUAU .(((((((((((((((((.-----..)))).))))))).))))))(((....))).(((((((-.(((((....)).))).))))))).((((((.----....))-----)))) ( -38.90) >DroSec_CAF1 360 109 + 1 UCCCCAACACGUCAGCAGA-----AUCUGUGUGAUGUGCUUGGGAGGAAAUUUCCAAGCGGCA-AGAUGUGAUAACGAUUGUGUCGCUGAUAUGUUAGAUAUUAGCAUUAGAUAU ..((((((((((((((((.-----..)))).))))))).))))).(((....)))....((((-.(((((....)).))).))))((((((((.....))))))))......... ( -34.60) >DroEre_CAF1 267024 97 + 1 UCCCCAACACGUCAGCAGAAUCGAAUCUGUAUAAUGUGCGCGGGGGG-AGUUUCCAAGCGGCAAAGAUGUGAUAAAGAUCGUGUUAUUGAUAUGUA----CA------------- .((((..(((((..(((((......)))))...)))))...))))((-.....))..(((.(((.((..((((....))))..)).)))...))).----..------------- ( -25.70) >DroYak_CAF1 79287 98 + 1 UCCCCAACACGUCAGCAGA-----AUCUGUAUGAUGUGCUCGGGGGG-AAUUUCCGAGCGGCA-AGAUGUGAUAAAGAUCGUGUUACUGAUAUGCA----CUUAG------AUGU (((((..(((((((((((.-----..)))).))))))).....))))-)...((..((..(((-....((((((.......)))))).....))).----))..)------)... ( -27.40) >consensus UCCCCAACACGUCAGCAGA_____AUCUGUAUGAUGUGCUCGGGAGG_AAUUUCCAAGCGGCA_AGAUGUGAUAAAGAUCGUGUCACUGAUAUGUA____CUUAG______AUAU ........(((((((((((......))))).))))))(((((((((....))))))))).(((.....((((((.......)))))).....))).................... (-26.25 = -25.62 + -0.62)

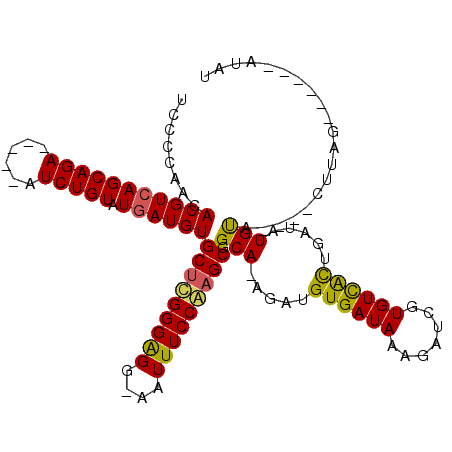

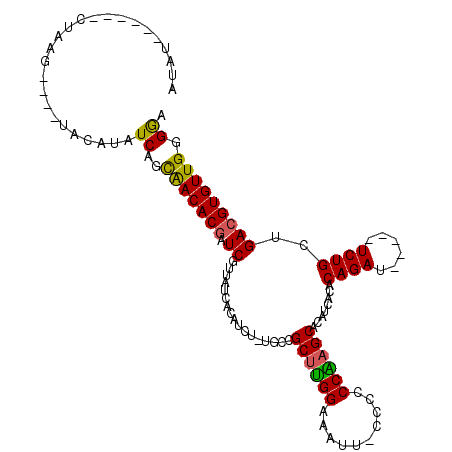
| Location | 21,648,736 – 21,648,836 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 78.54 |
| Mean single sequence MFE | -23.18 |
| Consensus MFE | -19.88 |
| Energy contribution | -19.25 |
| Covariance contribution | -0.62 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.631495 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21648736 100 - 22224390 AUAU-----GCCAAG----UACAUAUCAGCGACACGAUCGUUAUCACAUCU-UGCCGCUUGGAAAUUUCCUCCCAAGCACAUCACACAGAU-----UCUGCUGACGUGUUGGGAU ..((-----.(((..----.(((..((((((.((.(((.((....))))).-)).)((((((..........)))))).............-----...)))))..)))))).)) ( -24.20) >DroSec_CAF1 360 109 - 1 AUAUCUAAUGCUAAUAUCUAACAUAUCAGCGACACAAUCGUUAUCACAUCU-UGCCGCUUGGAAAUUUCCUCCCAAGCACAUCACACAGAU-----UCUGCUGACGUGUUGGGGA ................(((((((..((((((((......))).........-....((((((..........)))))).............-----...)))))..))))))).. ( -26.30) >DroEre_CAF1 267024 97 - 1 -------------UG----UACAUAUCAAUAACACGAUCUUUAUCACAUCUUUGCCGCUUGGAAACU-CCCCCCGCGCACAUUAUACAGAUUCGAUUCUGCUGACGUGUUGGGGA -------------..----......((..(((((((.((.............(((((...((.....-))...)).))).......((((......))))..)))))))))..)) ( -19.20) >DroYak_CAF1 79287 98 - 1 ACAU------CUAAG----UGCAUAUCAGUAACACGAUCUUUAUCACAUCU-UGCCGCUCGGAAAUU-CCCCCCGAGCACAUCAUACAGAU-----UCUGCUGACGUGUUGGGGA ...(------((...----.(((..((((((....(((((.(((.......-((..((((((.....-....)))))).))..))).))))-----).))))))..)))...))) ( -23.00) >consensus AUAU______CUAAG____UACAUAUCAGCAACACGAUCGUUAUCACAUCU_UGCCGCUUGGAAAUU_CCCCCCAAGCACAUCACACAGAU_____UCUGCUGACGUGUUGGGGA .........................((..(((((((.((.................((((((..........))))))........((((......))))..))))))))).)). (-19.88 = -19.25 + -0.62)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:08:41 2006