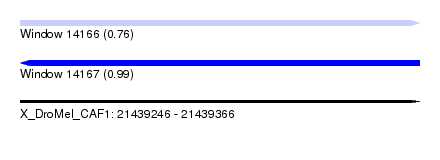
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 21,439,246 – 21,439,366 |
| Length | 120 |
| Max. P | 0.987023 |

| Location | 21,439,246 – 21,439,366 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 61.82 |
| Mean single sequence MFE | -41.23 |
| Consensus MFE | -19.20 |
| Energy contribution | -19.53 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.755762 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21439246 120 + 22224390 UGGCCAGUUGAACGGGGGCAGGAGCCCCUGGCGACUGAACAGCCAAAAAUGCGUCCUGUGGCGAUUGAACAGCCACCGGGGAGGCCGAUGUCGGAUAUAAAGCCACCGGAGAAGACUGCU ((((((((((..(((((((....))))))).))))))....)))).....(((((...((((.........))))((((((...(((....)))........)).))))....))).)). ( -47.30) >DroSim_CAF1 6763 90 + 1 UCGGCACUUGAACGGGGACAGGAGUCGAUGGCUGCUGAACAGCCACCGGAGUGGCCGGUGUCGGAUAUGAAGCCACCGG------------------------------AGGAGACUGCU ..(((.......(((((.((....((((((((((.....)))))(((((.....))))))))))...))...)).))).------------------------------((....))))) ( -32.80) >DroYak_CAF1 13060 90 + 1 UCGCCACUUGAACGGGGACAGGAGCCGGUGGCGACUGAACAGCCACCGGGGCAGCCGGUGCCUGAAAUUCAGCCACCGG------------------------------AAGAGCCUCCU ((.((........)).)).((((.((((((((.........))))))))(((..((((((.(((.....))).))))))------------------------------....))))))) ( -43.60) >consensus UCGCCACUUGAACGGGGACAGGAGCCGAUGGCGACUGAACAGCCACCGGAGCGGCCGGUGCCGGAUAUACAGCCACCGG______________________________AGGAGACUGCU ............(((((.(((..(((...)))..)))....(.((((((.....)))))).)..........)).))).......................................... (-19.20 = -19.53 + 0.34)

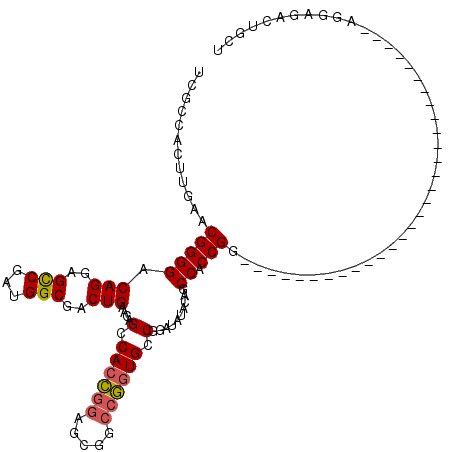
| Location | 21,439,246 – 21,439,366 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 61.82 |
| Mean single sequence MFE | -38.87 |
| Consensus MFE | -20.75 |
| Energy contribution | -20.20 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.53 |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.987023 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21439246 120 - 22224390 AGCAGUCUUCUCCGGUGGCUUUAUAUCCGACAUCGGCCUCCCCGGUGGCUGUUCAAUCGCCACAGGACGCAUUUUUGGCUGUUCAGUCGCCAGGGGCUCCUGCCCCCGUUCAACUGGCCA ..((((.......(((((((..(((.((((.....((.(((...(((((.........))))).))).))....)))).)))..))))))).(((((....)))))......)))).... ( -44.50) >DroSim_CAF1 6763 90 - 1 AGCAGUCUCCU------------------------------CCGGUGGCUUCAUAUCCGACACCGGCCACUCCGGUGGCUGUUCAGCAGCCAUCGACUCCUGUCCCCGUUCAAGUGCCGA .(.((((.((.------------------------------..)).)))).)...........((((.(((..((((((((.....))))))))(((....)))........))))))). ( -29.80) >DroYak_CAF1 13060 90 - 1 AGGAGGCUCUU------------------------------CCGGUGGCUGAAUUUCAGGCACCGGCUGCCCCGGUGGCUGUUCAGUCGCCACCGGCUCCUGUCCCCGUUCAAGUGGCGA (((((((....------------------------------((((((.(((.....))).))))))..)))((((((((.........)))))))).)))).((.(((......))).)) ( -42.30) >consensus AGCAGUCUCCU______________________________CCGGUGGCUGAAUAUCCGACACCGGCCGCACCGGUGGCUGUUCAGUCGCCACCGGCUCCUGUCCCCGUUCAAGUGGCGA .((((....................................((((((.(.........).)))))).....((((((((.........))))))))...))))................. (-20.75 = -20.20 + -0.55)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:08:39 2006