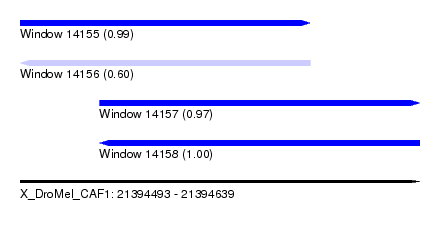
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 21,394,493 – 21,394,639 |
| Length | 146 |
| Max. P | 0.996575 |

| Location | 21,394,493 – 21,394,599 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 82.26 |
| Mean single sequence MFE | -28.12 |
| Consensus MFE | -22.76 |
| Energy contribution | -23.82 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.81 |
| SVM decision value | 2.35 |
| SVM RNA-class probability | 0.992781 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21394493 106 + 22224390 ---A--AAAAA----ACUCGAGGAAAAAACUUAAAUCUGUUGACUGGACAGCGGAGGCAAUGAUUUAGUGGCCUAUAAAAGGCCACUUCCCGGACCUA-AAACCCCAACCAACGAU ---.--.....----..(((.((((..........(((((((......)))))))............(((((((.....)))))))))))))).....-................. ( -26.30) >DroSec_CAF1 42725 100 + 1 --------AAA----AACCGAGGAAAAAACUGAAAGCUGUUGACUGGACAGCGGAGGCAAUGAUUUAGUGGCCUAUAAAAGGCCACUUCCCUGACCUA-AAACCCCAACCA---AU --------...----......((............((((((.....))))))..(((((..((...((((((((.....))))))))))..)).))).-.........)).---.. ( -27.30) >DroEre_CAF1 30540 106 + 1 CAACAGCAAAA----AAGCGAGGCAAAAA--GAAAUCUGUUGACUGGACAGCGGAGGCAGUGAUUUAGUGGCCUAUAAAAGGCCACUUCCCUGAACUA-AAGCCCCAACCA---AU .....((....----..))(.(((.....--....(((((((......)))))))..(((.((...((((((((.....)))))))))).))).....-..))).).....---.. ( -31.10) >DroYak_CAF1 34647 111 + 1 AAACAGAAAAAGAAUAACCGUGGCAAAGA--GGAAUCUGUUGACUGGACAGCGGAGGCAGUGAGUUAGUGGCCUAUUAAAGGUCACUUCCCUGACCAAAAAACCCCAACCA---AU ....................(((....(.--((..(((((((......)))))))(((((.((...((((((((.....)))))))))).))).))......)))...)))---.. ( -27.80) >consensus ___C__AAAAA____AACCGAGGAAAAAA__GAAAUCUGUUGACUGGACAGCGGAGGCAAUGAUUUAGUGGCCUAUAAAAGGCCACUUCCCUGACCUA_AAACCCCAACCA___AU .....................((............(((((((......)))))))(((((.((...((((((((.....)))))))))).))).))............))...... (-22.76 = -23.82 + 1.06)

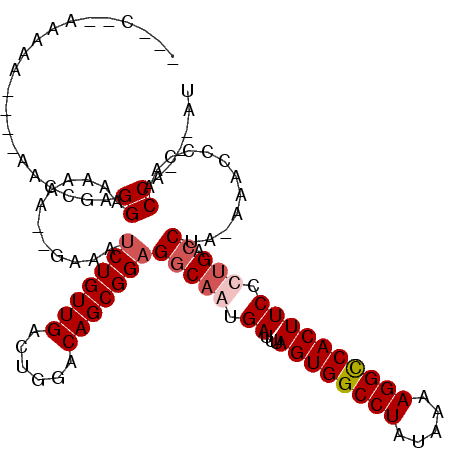

| Location | 21,394,493 – 21,394,599 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 82.26 |
| Mean single sequence MFE | -29.05 |
| Consensus MFE | -21.69 |
| Energy contribution | -22.25 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.598164 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21394493 106 - 22224390 AUCGUUGGUUGGGGUUU-UAGGUCCGGGAAGUGGCCUUUUAUAGGCCACUAAAUCAUUGCCUCCGCUGUCCAGUCAACAGAUUUAAGUUUUUUCCUCGAGU----UUUUU--U--- ...((((..((((((..-.((((..((..((((((((.....))))))))...))...))))..)))..)))..))))(((((..((.......))..)))----))...--.--- ( -31.30) >DroSec_CAF1 42725 100 - 1 AU---UGGUUGGGGUUU-UAGGUCAGGGAAGUGGCCUUUUAUAGGCCACUAAAUCAUUGCCUCCGCUGUCCAGUCAACAGCUUUCAGUUUUUUCCUCGGUU----UUU-------- ..---...((((((...-.(((.(((.((((((((((.....))))))))...)).))))))..(((((.......)))))............))))))..----...-------- ( -29.10) >DroEre_CAF1 30540 106 - 1 AU---UGGUUGGGGCUU-UAGUUCAGGGAAGUGGCCUUUUAUAGGCCACUAAAUCACUGCCUCCGCUGUCCAGUCAACAGAUUUC--UUUUUGCCUCGCUU----UUUUGCUGUUG ..---.....(((((..-..(..((((((((((((((.....))))))))...........))).)))..)((((....))))..--.....)))))((..----....))..... ( -30.00) >DroYak_CAF1 34647 111 - 1 AU---UGGUUGGGGUUUUUUGGUCAGGGAAGUGACCUUUAAUAGGCCACUAACUCACUGCCUCCGCUGUCCAGUCAACAGAUUCC--UCUUUGCCACGGUUAUUCUUUUUCUGUUU ..---.(((.((((..((((((.((((((((((.(((.....))).))))...........))).))).)))......)))..))--))...)))((((..(.....)..)))).. ( -25.80) >consensus AU___UGGUUGGGGUUU_UAGGUCAGGGAAGUGGCCUUUUAUAGGCCACUAAAUCACUGCCUCCGCUGUCCAGUCAACAGAUUUC__UUUUUGCCUCGGUU____UUUUU__G___ .....((..((((((....(((.(((.((((((((((.....))))))))...)).))))))..)))..)))..))........................................ (-21.69 = -22.25 + 0.56)



| Location | 21,394,522 – 21,394,639 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.72 |
| Mean single sequence MFE | -32.46 |
| Consensus MFE | -17.45 |
| Energy contribution | -20.77 |
| Covariance contribution | 3.32 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.974177 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21394522 117 + 22224390 GUUGACUGGACAGCGGAGGCAAUGAUUUAGUGGCCUAUAAAAGGCCACUUC--CCGGACCUA-AAACCCCAACCAACGAUCUUAAAAAGGAGAAACCCCCGAUGGAAGUCGUAAAAGUUU ...((((...((.(((((((...((...((((((((.....))))))))))--...).))).-................((((......)))).....))).))..)))).......... ( -29.00) >DroSec_CAF1 42751 114 + 1 GUUGACUGGACAGCGGAGGCAAUGAUUUAGUGGCCUAUAAAAGGCCACUUC--CCUGACCUA-AAACCCCAACCA---AUCUUAAAAAGGGGAAGCCCCCGAUGGAAGUCGUAAAAGUUU ...((((...((.(((.(((........((((((((.....))))))))..--.........-...((((.....---..........))))..))).))).))..)))).......... ( -37.26) >DroEre_CAF1 30572 113 + 1 GUUGACUGGACAGCGGAGGCAGUGAUUUAGUGGCCUAUAAAAGGCCACUUC--CCUGAACUA-AAGCCCCAACCA---AUCUUGAAAAGGGGAAGCC-CCGCUGGAAGUCGUAAAAGUUU ...((((...((((((.((((((.....((((((((.....))))))))..--.....))).-...((((.....---..........))))..)))-))))))..)))).......... ( -45.06) >DroYak_CAF1 34683 115 + 1 GUUGACUGGACAGCGGAGGCAGUGAGUUAGUGGCCUAUUAAAGGUCACUUC--CCUGACCAAAAAACCCCAACCA---AUCUUAAAAAAGGGAAGCCCCCGAUGGAAGUCGUAAAAGUCU ...((((...((.(((.(((.((.((..((((((((.....))))))))..--.)).))........(((.....---...........)))..))).))).))..)))).......... ( -34.59) >DroAna_CAF1 37720 92 + 1 -----CCAGAUAGCAAUGUCAGUGAUUUUGUGGCCCAUAAAUGCCAACUUUGGCCUGACCCA-AAAA----------------AAAAAGUACAAA------AUAUAUGUUACGAGGAUGU -----((.((((....))))....((((((((((........)))...(((((......)))-))..----------------.......)))))------))...........)).... ( -16.40) >consensus GUUGACUGGACAGCGGAGGCAGUGAUUUAGUGGCCUAUAAAAGGCCACUUC__CCUGACCUA_AAACCCCAACCA___AUCUUAAAAAGGGGAAGCCCCCGAUGGAAGUCGUAAAAGUUU ...((((...((.(((.(((.........(((((((.....)))))))..................((((..................))))..))).))).))..)))).......... (-17.45 = -20.77 + 3.32)

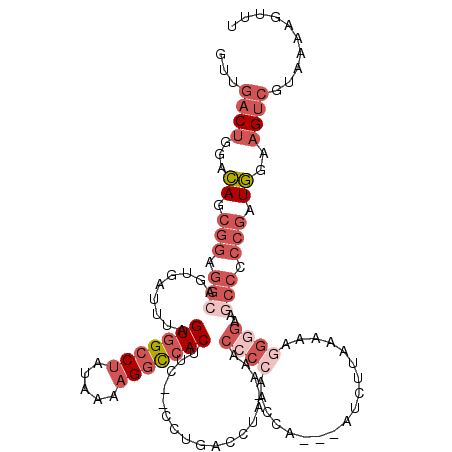

| Location | 21,394,522 – 21,394,639 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.72 |
| Mean single sequence MFE | -35.42 |
| Consensus MFE | -21.03 |
| Energy contribution | -23.51 |
| Covariance contribution | 2.48 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.59 |
| SVM decision value | 2.72 |
| SVM RNA-class probability | 0.996575 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21394522 117 - 22224390 AAACUUUUACGACUUCCAUCGGGGGUUUCUCCUUUUUAAGAUCGUUGGUUGGGGUUU-UAGGUCCGG--GAAGUGGCCUUUUAUAGGCCACUAAAUCAUUGCCUCCGCUGUCCAGUCAAC ..........((((..((.(((((((...((((.(((((((((.(.....).)))))-))))...))--))((((((((.....))))))))........))))))).))...))))... ( -40.30) >DroSec_CAF1 42751 114 - 1 AAACUUUUACGACUUCCAUCGGGGGCUUCCCCUUUUUAAGAU---UGGUUGGGGUUU-UAGGUCAGG--GAAGUGGCCUUUUAUAGGCCACUAAAUCAUUGCCUCCGCUGUCCAGUCAAC ..........((((..((.(((((((.((((...((((((((---(......)))))-))))...))--))((((((((.....))))))))........))))))).))...))))... ( -43.00) >DroEre_CAF1 30572 113 - 1 AAACUUUUACGACUUCCAGCGG-GGCUUCCCCUUUUCAAGAU---UGGUUGGGGCUU-UAGUUCAGG--GAAGUGGCCUUUUAUAGGCCACUAAAUCACUGCCUCCGCUGUCCAGUCAAC ..........((((..((((((-(((..((((...(((....---)))..))))...-.......((--..((((((((.....))))))))...))...))).))))))...))))... ( -43.40) >DroYak_CAF1 34683 115 - 1 AGACUUUUACGACUUCCAUCGGGGGCUUCCCUUUUUUAAGAU---UGGUUGGGGUUUUUUGGUCAGG--GAAGUGACCUUUAAUAGGCCACUAACUCACUGCCUCCGCUGUCCAGUCAAC ..........((((..((.(((((((....(((....)))..---.(((((((((((...(((((..--....)))))......))))).))))))....))))))).))...))))... ( -35.00) >DroAna_CAF1 37720 92 - 1 ACAUCCUCGUAACAUAUAU------UUUGUACUUUUU----------------UUUU-UGGGUCAGGCCAAAGUUGGCAUUUAUGGGCCACAAAAUCACUGACAUUGCUAUCUGG----- ............((.((((------..(((......(----------------(((.-(((.(((.((((....)))).....))).))).))))......)))..).))).)).----- ( -15.40) >consensus AAACUUUUACGACUUCCAUCGGGGGCUUCCCCUUUUUAAGAU___UGGUUGGGGUUU_UAGGUCAGG__GAAGUGGCCUUUUAUAGGCCACUAAAUCACUGCCUCCGCUGUCCAGUCAAC ..........((((..((.(((((((..((((..................)))).................((((((((.....))))))))........))))))).))...))))... (-21.03 = -23.51 + 2.48)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:08:31 2006