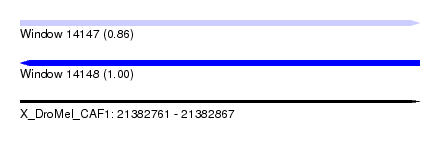
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 21,382,761 – 21,382,867 |
| Length | 106 |
| Max. P | 0.995759 |

| Location | 21,382,761 – 21,382,867 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 62.05 |
| Mean single sequence MFE | -19.65 |
| Consensus MFE | -8.80 |
| Energy contribution | -8.96 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.856148 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21382761 106 + 22224390 GUGUAAAAAACUCCGCACAGGUCUUGAAGGCAUUGCCAAAU--UUGU--AAAAUUACUGAGCUCUACUUUCCAGAUUAUUAUCAUUUCCAUUUUUCUUCCAUUCU-------AUUAU ((((..........)))).(((((.(((((....(((((((--(...--..))))..)).))....))))).)))))............................-------..... ( -11.30) >DroSec_CAF1 28459 98 + 1 GUGUAAAUAACUCCGCACAGGUCUUGAAUGCAUUGCCAAAU--UUGU--AAAAUUACUGAGCUCUACUUUCUAGACCUUGAGGGAA-------UACUUC-ACUAC-------AAAAA ((((..........))))((((((.(((......(((((((--(...--..))))..)).))......))).))))))(((((...-------..))))-)....-------..... ( -17.50) >DroEre_CAF1 19281 93 + 1 CUUUGAAAAACUCCGCAAAGGUCUAGAAUGUAUUGCUAAAU--UCGC--AAA-UCACUGAGCUAUACUUUCCUGACCUUAAGGGAA-------U--UUC-ACAAC-------A--AU ...(((((...(((...((((((..(((.((((.(((....--....--...-......))).)))).)))..))))))..)))..-------)--)))-)....-------.--.. ( -21.03) >DroYak_CAF1 19475 96 + 1 AUGUGAGAAACUCCGCUAAGGUCUUAAAUGUAUUGCCAAAU--UUGU--AAAGCCACUAAGCUAUACAUUUCUGACCUUAAGGGAA-------UAAUUC-ACAAA-------G--AC .((((((....(((..(((((((..(((((((((((.....--..))--))(((......))).)))))))..)))))))..))).-------...)))-)))..-------.--.. ( -27.50) >DroAna_CAF1 28188 104 + 1 C---CAACACUUUCGCAAAGGUCUCGAAAACAUUCCACAAUAUUUCUAUUAGGGCACUGAGG--AACUUUCACGACCUUUACAAAG-------UGUUUC-GUCACGAAUUCUAUUCU .---.((((((((.(.(((((((..((((...((((.((....(((.....)))...)).))--)).))))..))))))).)))))-------))))..-................. ( -20.90) >consensus CUGUAAAAAACUCCGCAAAGGUCUUGAAUGCAUUGCCAAAU__UUGU__AAAAUCACUGAGCUAUACUUUCCAGACCUUAAGGGAA_______UACUUC_ACAAC_______A__AU ...........(((...((((((..(((((....((........................))....)))))..))))))..)))................................. ( -8.80 = -8.96 + 0.16)



| Location | 21,382,761 – 21,382,867 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 62.05 |
| Mean single sequence MFE | -27.23 |
| Consensus MFE | -12.38 |
| Energy contribution | -12.74 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.39 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.45 |
| SVM decision value | 2.61 |
| SVM RNA-class probability | 0.995759 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21382761 106 - 22224390 AUAAU-------AGAAUGGAAGAAAAAUGGAAAUGAUAAUAAUCUGGAAAGUAGAGCUCAGUAAUUUU--ACAA--AUUUGGCAAUGCCUUCAAGACCUGUGCGGAGUUUUUUACAC .....-------.........(((((((..............(((.(((.(((..((.(((.......--....--..)))))..))).))).)))((.....)).))))))).... ( -11.92) >DroSec_CAF1 28459 98 - 1 UUUUU-------GUAGU-GAAGUA-------UUCCCUCAAGGUCUAGAAAGUAGAGCUCAGUAAUUUU--ACAA--AUUUGGCAAUGCAUUCAAGACCUGUGCGGAGUUAUUUACAC .....-------...((-(((..(-------(((((...((((((.(((.(((..((.(((.......--....--..)))))..))).))).))))))..).)))))..))))).. ( -26.62) >DroEre_CAF1 19281 93 - 1 AU--U-------GUUGU-GAA--A-------UUCCCUUAAGGUCAGGAAAGUAUAGCUCAGUGA-UUU--GCGA--AUUUAGCAAUACAUUCUAGACCUUUGCGGAGUUUUUCAAAG ..--.-------.(((.-(((--(-------((((...((((((.((((.((((.(((.(((..-...--....--))).))).)))).)))).))))))...)))))))).))).. ( -29.60) >DroYak_CAF1 19475 96 - 1 GU--C-------UUUGU-GAAUUA-------UUCCCUUAAGGUCAGAAAUGUAUAGCUUAGUGGCUUU--ACAA--AUUUGGCAAUACAUUUAAGACCUUAGCGGAGUUUCUCACAU ..--.-------..(((-((...(-------((((.((((((((..((((((((.(((...((.....--.)).--....))).))))))))..)))))))).)))))...))))). ( -35.00) >DroAna_CAF1 28188 104 - 1 AGAAUAGAAUUCGUGAC-GAAACA-------CUUUGUAAAGGUCGUGAAAGUU--CCUCAGUGCCCUAAUAGAAAUAUUGUGGAAUGUUUUCGAGACCUUUGCGAAAGUGUUG---G .((((...)))).....-..((((-------(((((((((((((.((((((((--((.(((((............))))).)))))..))))).))))))))).)))))))).---. ( -33.00) >consensus AU__U_______GUAGU_GAAGUA_______UUCCCUAAAGGUCAGGAAAGUAGAGCUCAGUGACUUU__ACAA__AUUUGGCAAUGCAUUCAAGACCUUUGCGGAGUUUUUCACAC ................................(((.((((((((..(((.(((..(((.(((..............))).)))..))).)))..)))))))).)))........... (-12.38 = -12.74 + 0.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:08:19 2006