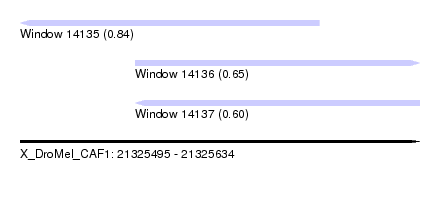
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 21,325,495 – 21,325,634 |
| Length | 139 |
| Max. P | 0.843285 |

| Location | 21,325,495 – 21,325,599 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.08 |
| Mean single sequence MFE | -36.43 |
| Consensus MFE | -28.72 |
| Energy contribution | -29.17 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.843285 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21325495 104 - 22224390 UGCCCUCUUUUCCAUUGUGAAAGGACAAAGGAGACGAAGGGCAGGCCAG----------------UAUUUGCAUCGGAUGAUCUGGCAAAUGUGAGGAAUAAUCCAGCGUAUUAUUCAAA (((((((.(((((.((((......)))).))))).).)))))).(((((----------------((((((...))))))..)))))..((((..(((....))).)))).......... ( -32.30) >DroEre_CAF1 64130 112 - 1 UGCCCUCUUUUCCAUUGUGACAGGACAAAGGAGUCGACGGGCAGCCCAGAGGCCGAG--------CAUUUGCAUCGGAUGAUCUGGCAAAUGUGAGGAGUAAUCCAGCGCAUUACUCAAA (((((((..((((.((((......)))).))))..)).)))))(((.(((..(((((--------(....)).))))....)))))).((((((.(((....)))..))))))....... ( -42.80) >DroYak_CAF1 67819 120 - 1 UGCCCUCUUUUCCAUUGUGAAAGGACAAAGGAGUCGAAGGGCAGCCCAGAGGCCAAGUAUAUAAGUAUUUGCAUCGGAUGAUCUGGCAAAUGUGAGGAAUAAUCCAGCGCAUUACUCAAA (((((((..((((.((((......)))).))))..).))))))(((.(((.((.((((((....)))))))).........)))))).((((((.(((....)))..))))))....... ( -34.20) >consensus UGCCCUCUUUUCCAUUGUGAAAGGACAAAGGAGUCGAAGGGCAGCCCAGAGGCC_AG________UAUUUGCAUCGGAUGAUCUGGCAAAUGUGAGGAAUAAUCCAGCGCAUUACUCAAA (((((((..((((.((((......)))).))))..)).))))).....(((..............(((((((..(((.....))))))))))...(((....))).........)))... (-28.72 = -29.17 + 0.45)

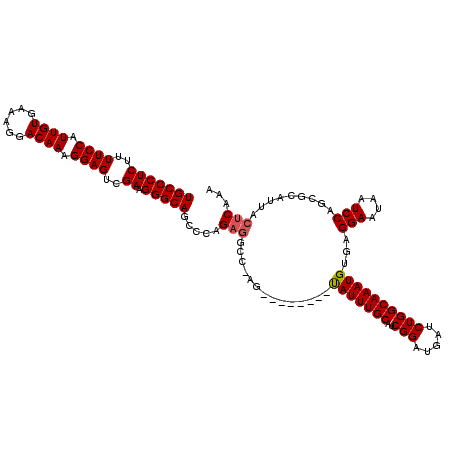
| Location | 21,325,535 – 21,325,634 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 86.65 |
| Mean single sequence MFE | -24.17 |
| Consensus MFE | -19.80 |
| Energy contribution | -20.80 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.649143 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21325535 99 + 22224390 CAUCCGAUGCAAAUA----------------CUGGCCUGCCCUUCGUCUCCUUUGUCCUUUCACAAUGGAAAAGAGGGCAUUUCAAAUCCAAUUUACUUAGUCAUCCCACUCAAA .....(((((.....----------------.(((..(((((((..(.(((.((((......)))).))))..)))))))..)))((......)).....).))))......... ( -18.50) >DroEre_CAF1 64170 107 + 1 CAUCCGAUGCAAAUG--------CUCGGCCUCUGGGCUGCCCGUCGACUCCUUUGUCCUGUCACAAUGGAAAAGAGGGCAUUUCAAAUCCAAUUUACUUAGUCAUCGCACUCAAA ....((((((....(--------(((((...))))))(((((.((...(((.((((......)))).)))...)))))))....................).)))))........ ( -27.50) >DroYak_CAF1 67859 115 + 1 CAUCCGAUGCAAAUACUUAUAUACUUGGCCUCUGGGCUGCCCUUCGACUCCUUUGUCCUUUCACAAUGGAAAAGAGGGCAUUUCAAAUCCAAUUUACUUAGUCAUCGCACUCAAA ....((((((................((((....))))((((((....(((.((((......)))).)))...)))))).....................).)))))........ ( -26.50) >consensus CAUCCGAUGCAAAUA________CU_GGCCUCUGGGCUGCCCUUCGACUCCUUUGUCCUUUCACAAUGGAAAAGAGGGCAUUUCAAAUCCAAUUUACUUAGUCAUCGCACUCAAA .....(((((.................(((....)))(((((.((...(((.((((......)))).)))...)))))))....................).))))......... (-19.80 = -20.80 + 1.00)



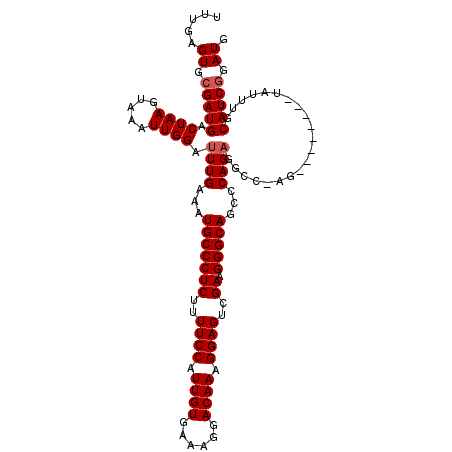
| Location | 21,325,535 – 21,325,634 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 86.65 |
| Mean single sequence MFE | -31.81 |
| Consensus MFE | -25.70 |
| Energy contribution | -26.37 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.597798 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21325535 99 - 22224390 UUUGAGUGGGAUGACUAAGUAAAUUGGAUUUGAAAUGCCCUCUUUUCCAUUGUGAAAGGACAAAGGAGACGAAGGGCAGGCCAG----------------UAUUUGCAUCGGAUG (((((.(((.....))).(((((((((........(((((((.(((((.((((......)))).))))).).))))))..))).----------------.)))))).))))).. ( -26.70) >DroEre_CAF1 64170 107 - 1 UUUGAGUGCGAUGACUAAGUAAAUUGGAUUUGAAAUGCCCUCUUUUCCAUUGUGACAGGACAAAGGAGUCGACGGGCAGCCCAGAGGCCGAG--------CAUUUGCAUCGGAUG .....((.(((((.(.((((...((((.((((...(((((((..((((.((((......)))).))))..)).)))))...))))..)))).--------.)))))))))).)). ( -33.80) >DroYak_CAF1 67859 115 - 1 UUUGAGUGCGAUGACUAAGUAAAUUGGAUUUGAAAUGCCCUCUUUUCCAUUGUGAAAGGACAAAGGAGUCGAAGGGCAGCCCAGAGGCCAAGUAUAUAAGUAUUUGCAUCGGAUG (((((.(((((..(((.......((((.((((...(((((((..((((.((((......)))).))))..).))))))...))))..)))).......)))..)))))))))).. ( -34.94) >consensus UUUGAGUGCGAUGACUAAGUAAAUUGGAUUUGAAAUGCCCUCUUUUCCAUUGUGAAAGGACAAAGGAGUCGAAGGGCAGCCCAGAGGCC_AG________UAUUUGCAUCGGAUG .....((.(((((.((((.....)))).((((...(((((((..((((.((((......)))).))))..)).)))))...)))).....................))))).)). (-25.70 = -26.37 + 0.67)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:08:08 2006