
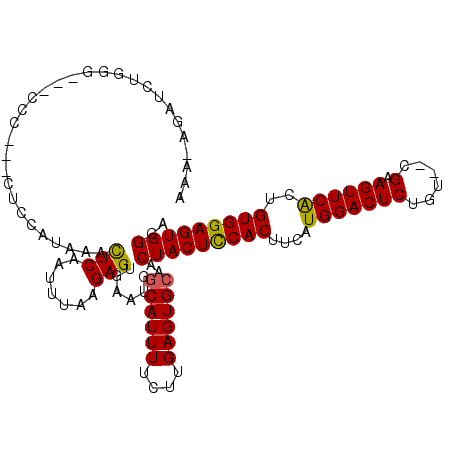
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 21,322,677 – 21,322,780 |
| Length | 103 |
| Max. P | 0.999773 |

| Location | 21,322,677 – 21,322,780 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 74.34 |
| Mean single sequence MFE | -36.53 |
| Consensus MFE | -22.93 |
| Energy contribution | -22.60 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.63 |
| SVM decision value | 4.05 |
| SVM RNA-class probability | 0.999773 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
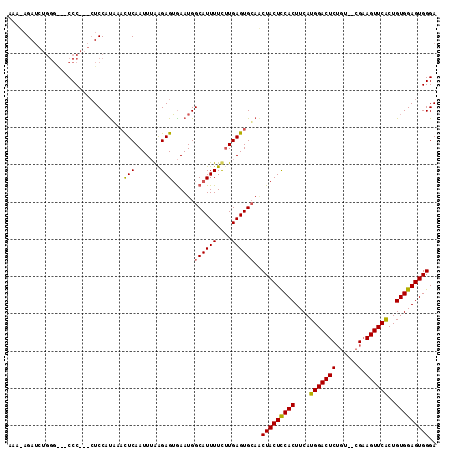
>X_DroMel_CAF1 21322677 103 + 22224390 -----GAUCUGGGA--CCCAAGAUCCUUGAACUCAAUUAAAGAGGAAAUGGCAUUUUUCUGAGUGCAACUACUCCACUUCAUGGACUC-----CGAAGUUCACUGUGGAGUGGGA -----(((((.(..--..).)))))......(((.......)))......((((((....))))))..(((((((((....((((((.-----...))))))..))))))))).. ( -32.60) >DroEre_CAF1 61361 112 + 1 AAACAGAUCUGGGGCUCCC---CUCCACAACUUCAAUUUGGGAGUGAAUGGCAUUUUCUUGAGUGCAGCUACUCCACUUCAUGGACUCUGUAGCGCAGUUCGCUGUGGAGUGGGA ...............((((---((((((((((((......)))))((((.((((((....)))))).(((((((((.....))))....)))))...))))..))))))).)))) ( -38.80) >DroYak_CAF1 65048 102 + 1 AAAGAGA---------CCC---CUCCAUUCACUCAAUUUAAGAGUGAAUGGCAUUUUCUUGAGUGGACCUACUUCACUUCAUGGACUCUGUC-CGAAGUUCACUGUGGAGUGGGA .((((((---------...---..((((((((((.......))))))))))...)))))).......((((((((((....((((((.....-...))))))..)))))))))). ( -38.20) >consensus AAA_AGAUCUGGG___CCC___CUCCAUAAACUCAAUUUAAGAGUGAAUGGCAUUUUCUUGAGUGCAACUACUCCACUUCAUGGACUCUGU__CGAAGUUCACUGUGGAGUGGGA ...............................(((.......)))......((((((....))))))..(((((((((....(((((((......).))))))..))))))))).. (-22.93 = -22.60 + -0.33)

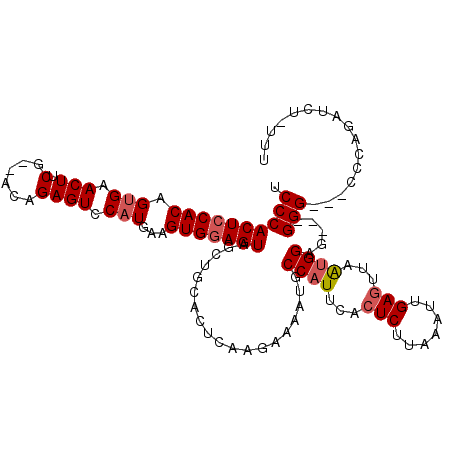
| Location | 21,322,677 – 21,322,780 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 74.34 |
| Mean single sequence MFE | -30.63 |
| Consensus MFE | -18.09 |
| Energy contribution | -19.20 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.633908 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21322677 103 - 22224390 UCCCACUCCACAGUGAACUUCG-----GAGUCCAUGAAGUGGAGUAGUUGCACUCAGAAAAAUGCCAUUUCCUCUUUAAUUGAGUUCAAGGAUCUUGGG--UCCCAGAUC----- .((((((((..((....))..)-----)))(((..(((((((.....(((....))).......)))))))(((.......))).....)))...))))--.........----- ( -23.10) >DroEre_CAF1 61361 112 - 1 UCCCACUCCACAGCGAACUGCGCUACAGAGUCCAUGAAGUGGAGUAGCUGCACUCAAGAAAAUGCCAUUCACUCCCAAAUUGAAGUUGUGGAG---GGGAGCCCCAGAUCUGUUU ((((.(((((((((((..((((((((....(((((...)))))))))).))).))............((((.........)))))))))))))---))))............... ( -35.90) >DroYak_CAF1 65048 102 - 1 UCCCACUCCACAGUGAACUUCG-GACAGAGUCCAUGAAGUGAAGUAGGUCCACUCAAGAAAAUGCCAUUCACUCUUAAAUUGAGUGAAUGGAG---GGG---------UCUCUUU ...((((....)))).((((((-(((...))))..)))))((((.((..((.(((..(......)(((((((((.......))))))))))))---)).---------.)))))) ( -32.90) >consensus UCCCACUCCACAGUGAACUUCG__ACAGAGUCCAUGAAGUGGAGUAGCUGCACUCAAGAAAAUGCCAUUCACUCUUAAAUUGAGUUAAUGGAG___GGG___CCCAGAUCU_UUU .((((((((((.(((.(((.(......)))).)))...)))))))...................((((...(((.......)))...)))).....)))................ (-18.09 = -19.20 + 1.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:08:05 2006