
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 21,318,297 – 21,318,447 |
| Length | 150 |
| Max. P | 0.848547 |

| Location | 21,318,297 – 21,318,416 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.83 |
| Mean single sequence MFE | -44.17 |
| Consensus MFE | -36.20 |
| Energy contribution | -36.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.750750 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
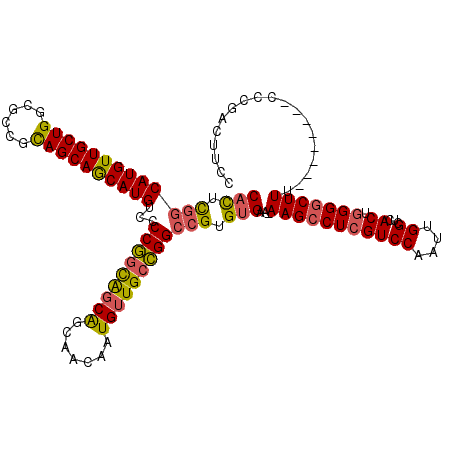
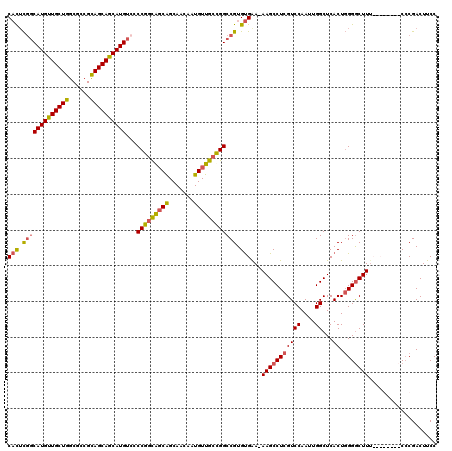
>X_DroMel_CAF1 21318297 119 - 22224390 CACUCGUCAUGUUGCUGGCGCCGCAGCAGCAUGUCGCCGGCAGCAGCAACAAUGUUGCCGGCCGUGUGAA-AAGCCUCGUCCAAUUGGCUCACUGGGGCUUUCCCGACUUCCCGACUUCC (((.((.((((((((((......)))))))))).)((((((((((.......)))))))))).).))).(-(((((((((((....))...)).)))))))).................. ( -50.00) >DroSec_CAF1 65086 119 - 1 CACUCGGCAUGUUGCUGGCGCCGCAGCAACAUGUCCCCGACAGCAGCAACAAUGUUGCCGGCCGUGUGGA-AAGCCUCGUCCAAUUGGCUCACUGGGGCUUUCCCGACUUCCCGACUUCC .....((((((((((((......)))))))))))).......((((((....))))))(((...((.(((-(((((((((((....))...)).)))))))))))).....)))...... ( -47.20) >DroEre_CAF1 57211 112 - 1 CACUCGGCAUGUUGCUGGCCUCGCAGCAGCAUGUGCCCGGCGUCAGCAACAAUGUUGCCGGCCGUGUGGAAAAGCCUCGUCCAAUUGGCUCACUGGGACUUU--------CACGACUUCC ...((((((((((((((......))))))))))).(((((((.(((((....))))).))((((..((((.........))))..))))...))))).....--------..)))..... ( -41.30) >DroYak_CAF1 60867 111 - 1 CUUGUUGCAUGUUGCUGGCGCCGUAGCAGCAUGUUCCCAGUGGCGGCAACAAUGUUGCUGGCCGUGCGAAAAAGCCUCGUCCAAUUGGCUCACUUGGGCUU---------CCCGACUUCC ...((((((((((((((......))))))))))..(((((((((((((((...))))))((.((.((......))..)).)).....)).))).))))...---------..)))).... ( -38.20) >consensus CACUCGGCAUGUUGCUGGCGCCGCAGCAGCAUGUCCCCGGCAGCAGCAACAAUGUUGCCGGCCGUGUGAA_AAGCCUCGUCCAAUUGGCUCACUGGGGCUUU________CCCGACUUCC (((.(((((((((((((......))))))))))...(((((((((.......)))))))))))).)))...(((((((((((....))...)).)))))))................... (-36.20 = -36.20 + -0.00)

| Location | 21,318,337 – 21,318,447 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 78.77 |
| Mean single sequence MFE | -44.41 |
| Consensus MFE | -34.56 |
| Energy contribution | -34.88 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.848547 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
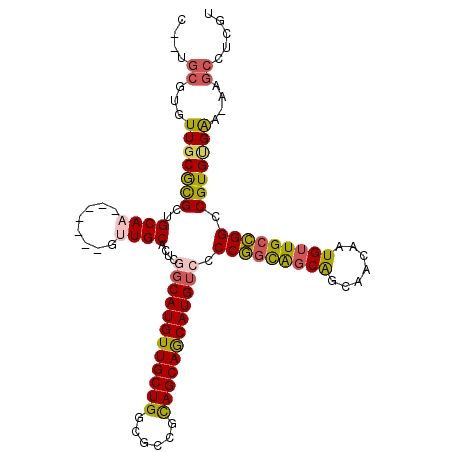
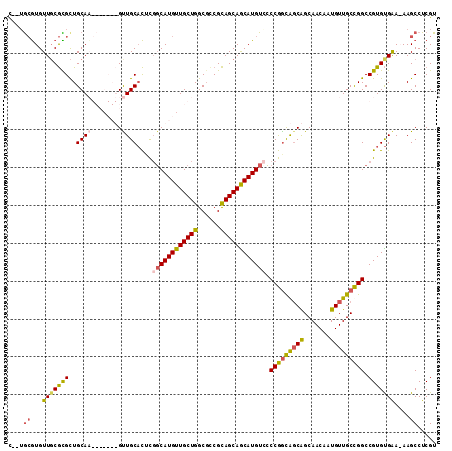
>X_DroMel_CAF1 21318337 110 - 22224390 U--UGCGUGUUGCGCGCUGCAAGUUGCAUGUUGCACUCGUCAUGUUGCUGGCGCCGCAGCAGCAUGUCGCCGGCAGCAGCAACAAUGUUGCCGGCCGUGUGAA-AAGCCUCGU (--((((((.....))).))))(..((...(..(((..(.((((((((((......)))))))))).)((((((((((.......)))))))))).)))..).-..))..).. ( -50.60) >DroSec_CAF1 65126 110 - 1 U--GGCGUGUUGCGCGCUGCAAGUUGCAUGUUGCACUCGGCAUGUUGCUGGCGCCGCAGCAACAUGUCCCCGACAGCAGCAACAAUGUUGCCGGCCGUGUGGA-AAGCCUCGU .--(((...(..((((((((((((((..((((((..((((((((((((((......))))))))))...))))..)))))).)))).))).))).))))..).-..))).... ( -49.00) >DroEre_CAF1 57243 104 - 1 C--UGCGUGUUUCGUGUUGCAU-------GUUGCACUCGGCAUGUUGCUGGCCUCGCAGCAGCAUGUGCCCGGCGUCAGCAACAAUGUUGCCGGCCGUGUGGAAAAGCCUCGU .--.((((((........))))-------))..(((.(((((((((((((......))))))))))...((((((.((.......)).))))))))).)))............ ( -38.30) >DroYak_CAF1 60898 106 - 1 CGUUAAAUGUUGCAUGCUGCAU-------GCUGCUUGUUGCAUGUUGCUGGCGCCGUAGCAGCAUGUUCCCAGUGGCGGCAACAAUGUUGCUGGCCGUGCGAAAAAGCCUCGU ((.......(((((((..((..-------((..((.(..(((((((((((......)))))))))))..).))..))((((((...)))))).)))))))))........)). ( -39.76) >consensus C__UGCGUGUUGCGCGCUGCAA_______GUUGCACUCGGCAUGUUGCUGGCGCCGCAGCAGCAUGUCCCCGGCAGCAGCAACAAUGUUGCCGGCCGUGUGAA_AAGCCUCGU ....((...(((((((..((((........))))....((((((((((((......)))))))))))).(((((((((.......))))))))).)))))))....))..... (-34.56 = -34.88 + 0.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:08:01 2006