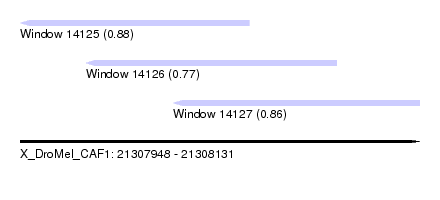
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 21,307,948 – 21,308,131 |
| Length | 183 |
| Max. P | 0.875423 |

| Location | 21,307,948 – 21,308,053 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 76.17 |
| Mean single sequence MFE | -25.05 |
| Consensus MFE | -12.46 |
| Energy contribution | -12.58 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.875423 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21307948 105 - 22224390 UCUGUGUGAGUGUGACCAUACAGACACAAGCGUA--CAUAUGUAUGUGUGUAACUCACAGAGAGAAAAAUUACCAUAGAACUCAGAGGUAUCUUAAAGAUACACUCA (((.((((((((((....)))..((((..(((((--(....)))))))))).))))))).))).....................((((((((.....))))).))). ( -31.10) >DroSec_CAF1 49452 93 - 1 UCUGUGUGAGUGUGACCAUAC------AAGCAUA--CAUA----UGUGUGUAACUCACAGAGAGAAAAAUCACCAUGGACUUCAGUGGUAUCUUGAAGAUA--CUCA ((((((.(.((((....))))------..(((((--(...----.))))))..).))))))(((.......(((((.(....).)))))(((.....))).--))). ( -24.80) >DroSim_CAF1 45892 93 - 1 UCUGUGUGAGUGUGACCAUAC------AAGCAUA--CAUA----UGUGUGUAACUCACAGAGAGAAAAAUCACCAUGGAAUUCAGUGGUAUCUUGAAGAUA--CUCA ((((((.(.((((....))))------..(((((--(...----.))))))..).))))))(((.......(((((.(....).)))))(((.....))).--))). ( -24.80) >DroEre_CAF1 48503 92 - 1 UUUGUGUGCGUGUGACCACAC------AGACACAAGCAUA----UGUGUGCACU-UGCAGUGAGAAAAAUUGCCGUCAAAUUUACUAUUUUUUU--AGUUA--CUUA ..((((((.(.....))))))------)(((....((((.----...))))...-.(((((.......))))).)))......((((......)--)))..--.... ( -19.50) >consensus UCUGUGUGAGUGUGACCAUAC______AAGCAUA__CAUA____UGUGUGUAACUCACAGAGAGAAAAAUCACCAUGGAAUUCAGUGGUAUCUUGAAGAUA__CUCA (((.(((((((.....(((((........................)))))..))))))).)))........(((((.(....).))))).................. (-12.46 = -12.58 + 0.12)

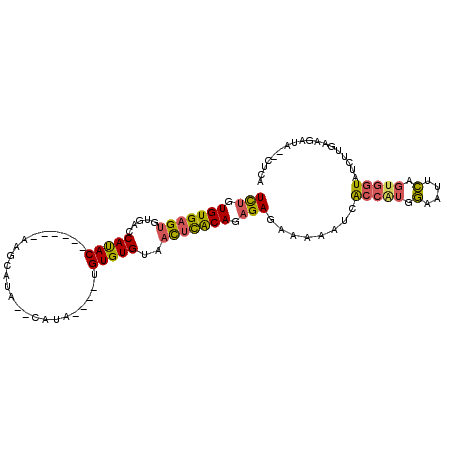
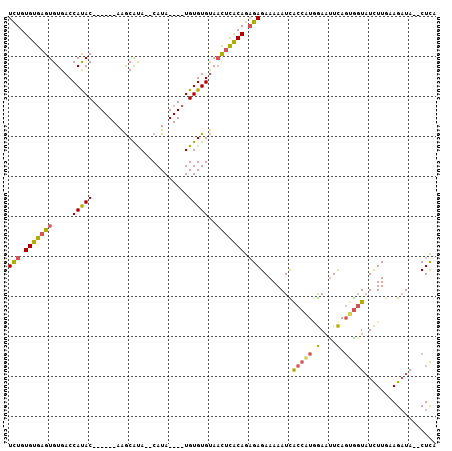
| Location | 21,307,978 – 21,308,093 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 86.04 |
| Mean single sequence MFE | -27.80 |
| Consensus MFE | -17.72 |
| Energy contribution | -17.76 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.770616 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21307978 115 - 22224390 AUGAUGGGAAUUUUACUAAGAAUUUUUCAUUUCCUUUAACUCUGUGUGAGUGUGACCAUACAGACACAAGCGUA--CAUAUGUAUGUGUGUAACUCACAGAGAGAAAAAUUACCAUA ...((((.((((((.....(((........)))......((((.((((((((((....)))..((((..(((((--(....)))))))))).))))))).)))).)))))).)))). ( -31.90) >DroSec_CAF1 49480 105 - 1 AUGAUGGGAAUUUUACUAAGAAUUUUUCAUUUCCUUUAACUCUGUGUGAGUGUGACCAUAC------AAGCAUA--CAUA----UGUGUGUAACUCACAGAGAGAAAAAUCACCAUG ...((((..(((((.....(((........)))......((((.((((((((((....)))------..(((((--(...----.)))))).))))))).)))).)))))..)))). ( -29.50) >DroSim_CAF1 45920 105 - 1 AUGAUGGGAAUUUUACUAAGAAUUUUUCAUUUCCUUUAACUCUGUGUGAGUGUGACCAUAC------AAGCAUA--CAUA----UGUGUGUAACUCACAGAGAGAAAAAUCACCAUG ...((((..(((((.....(((........)))......((((.((((((((((....)))------..(((((--(...----.)))))).))))))).)))).)))))..)))). ( -29.50) >DroEre_CAF1 48529 106 - 1 AUGAUGGGAAUUUUACUAAGAAUUUUUCAUUUCCUUUAACUUUGUGUGCGUGUGACCACAC------AGACACAAGCAUA----UGUGUGCACU-UGCAGUGAGAAAAAUUGCCGUC ..(((((.((((((..........(((((((..........((((((..((((....))))------..))))))(((..----((....))..-)))))))))))))))).))))) ( -27.10) >DroYak_CAF1 51079 106 - 1 AUGAUGGGAAUUUUACUAAGAAUUUUUCAUUUCCUUUAACUCUGUGUGAGUGUGACCAUAC------AGACACAAGCAUA----UGUGUGUGAC-UGCAGUGAGAAAAAAUACCUUG .....(((.(((((.........((((((((.........((((((((........)))))------)))((((.((...----.)).))))..-...))))))))))))).))).. ( -21.00) >consensus AUGAUGGGAAUUUUACUAAGAAUUUUUCAUUUCCUUUAACUCUGUGUGAGUGUGACCAUAC______AAGCAUA__CAUA____UGUGUGUAACUCACAGAGAGAAAAAUUACCAUG ..((((..((.(((.....))).))..))))........((((.(((((((.....(((((........................)))))..))))))).))))............. (-17.72 = -17.76 + 0.04)

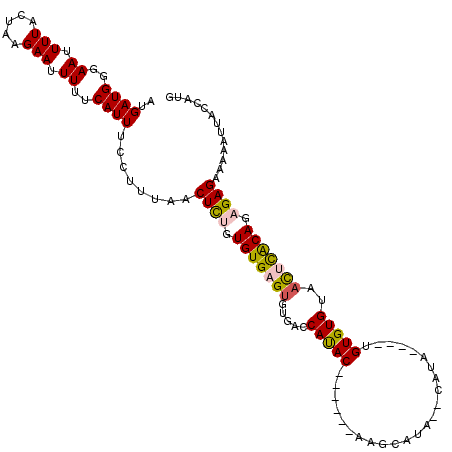
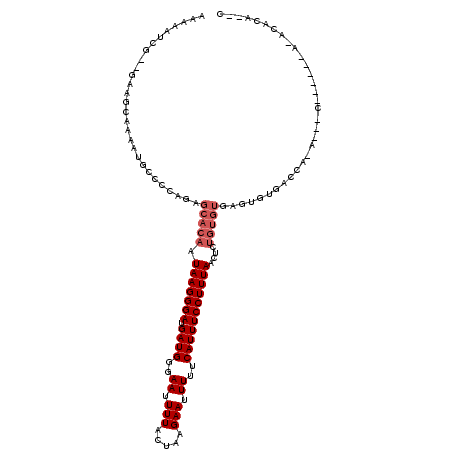
| Location | 21,308,018 – 21,308,131 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.03 |
| Mean single sequence MFE | -26.38 |
| Consensus MFE | -14.38 |
| Energy contribution | -15.22 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.856000 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21308018 113 - 22224390 AAAAAUCG--GGAGCAAAAUGCCCCAGAGCACAAUAAGGGAUGAUGGGAAUUUUACUAAGAAUUUUUCAUUUCCUUUAACUCUGUGUGAGUGUGACCAUA---CAGACACAAGCGUA--C .......(--((.((.....)))))...((....(((((((.((((..((.(((.....))).))..)))))))))))..((((((((........))))---)))).....))...--. ( -32.40) >DroPse_CAF1 99346 97 - 1 AAAAAACCAAAAGGCAAAAUGCCCCAGAGCACAAUAAGGGAUGAUGGGAAUUUUACUAAGAAUUUUUCAUUUCCUUUAACUUUGUGUGUGUG-GACGA---------------------- ............(((.....)))(((..(((((((((((((.((((..((.(((.....))).))..)))))))))))...))))))...))-)....---------------------- ( -25.90) >DroEre_CAF1 48564 107 - 1 GAAAAUCG--GG--CGAAAUGCCCCAGAGCACAAUAAGGGAUGAUGGGAAUUUUACUAAGAAUUUUUCAUUUCCUUUAACUUUGUGUGCGUGUGACCACA---C------AGACACAAGC .......(--((--(.....))))....((....(((((((.((((..((.(((.....))).))..)))))))))))...((((((..((((....)))---)------..)))))))) ( -30.70) >DroYak_CAF1 51114 109 - 1 AAAAAUCG--GGACCCAAAUGCCCCAGAGCACAAUAAGGGAUGAUGGGAAUUUUACUAAGAAUUUUUCAUUUCCUUUAACUCUGUGUGAGUGUGACCAUA---C------AGACACAAGC .......(--((..........)))...((....(((((((.((((..((.(((.....))).))..)))))))))))..((((((((........))))---)------))).....)) ( -26.60) >DroAna_CAF1 40345 103 - 1 AAAAAUUG--GAAGCAAAAUGCCCCAGAGCACAAUAAGGGAUGAUGGGAAUUUUACUAAGAAUUUUUCAUUUCCUUUAACACA-------CACGGCCACAGGAC------ACACACA--C .....(((--...((....(((......)))...(((((((.((((..((.(((.....))).))..))))))))))).....-------....))..)))...------.......--. ( -16.80) >DroPer_CAF1 75844 97 - 1 AAAAAACCAAAAGGCAAAAUGCCCCAGAGCACAAUAAGGGAUGAUGGGAAUUUUACUAAGAAUUUUUCAUUUCCUUUAACUUUGUGUGUGUG-GACGA---------------------- ............(((.....)))(((..(((((((((((((.((((..((.(((.....))).))..)))))))))))...))))))...))-)....---------------------- ( -25.90) >consensus AAAAAUCG__GAAGCAAAAUGCCCCAGAGCACAAUAAGGGAUGAUGGGAAUUUUACUAAGAAUUUUUCAUUUCCUUUAACUCUGUGUGAGUGUGACCA_A___C______A_ACACA__C ............................(((((.(((((((.((((..((.(((.....))).))..)))))))))))....)))))................................. (-14.38 = -15.22 + 0.83)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:07:59 2006