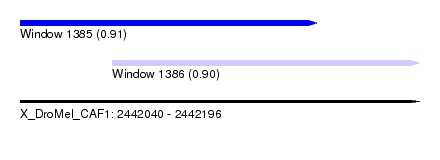
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,442,040 – 2,442,196 |
| Length | 156 |
| Max. P | 0.914806 |

| Location | 2,442,040 – 2,442,156 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.24 |
| Mean single sequence MFE | -38.27 |
| Consensus MFE | -24.24 |
| Energy contribution | -24.93 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.914806 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2442040 116 + 22224390 AAAAGCAAGGAAUCGAGUAAAC----ACGUUCCACCACCGCUGUGGCAAUCCGUGGCUGGUGCGCAACGGCCACACUGACGCGCUGACCAAUUUUUAGUUAUUAGUGUGACCUUAAGCAU ....((..((((.((.(....)----.))))))..((((((..(((....)))..)).)))).))...((.((((((((...(((((.......)))))..)))))))).))........ ( -35.80) >DroSec_CAF1 2942 111 + 1 -----CGGGGAACCGAGUAAAC----ACGUUCCACCACCGCUGUGGCAAUCCGCUGAUGGUGCGCAGCGGUCACAUUGACGCGCUGACCAAUUUUUAGUUAUUAGUGUGACCGUAUGCAU -----.(((((((.(.(....)----.)))))).))(((((.((((....)))).).))))..((((((((((((((((...(((((.......)))))..))))))))))))).))).. ( -41.20) >DroSim_CAF1 5095 111 + 1 -----CGGGCAACCGAGUAAAC----ACGUUCCACCACCGCUGUGGCAAUCCGCUGAUGGUGCGCAACGGUCACAUUGACGCGCUGACCAAUUUUUAGUUAUUAGUGUGACCGUUAGCAU -----(((....))).......----.........((((((.((((....)))).).))))).((((((((((((((((...(((((.......)))))..)))))))))))))).)).. ( -41.50) >DroYak_CAF1 3466 113 + 1 -----CAAUUAGCGCAUUAUAUAUCAAUAUUCCACCACCGCUGUGGCAAUCCGUUGCUUGUGCGCAACGGUCACACUGACG--CUGGGCACUUUGUAUUUAUUAGUGUGGCCGUAAUCAU -----......((((((.......((((......((((....))))......))))...)))))).(((((((((((((..--....((.....)).....)))))))))))))...... ( -34.60) >consensus _____CAAGGAACCGAGUAAAC____ACGUUCCACCACCGCUGUGGCAAUCCGCUGAUGGUGCGCAACGGUCACACUGACGCGCUGACCAAUUUUUAGUUAUUAGUGUGACCGUAAGCAU ...................................((((((.((((....)))).).)))))....(((((((((((((...(((((.......)))))..)))))))))))))...... (-24.24 = -24.93 + 0.69)

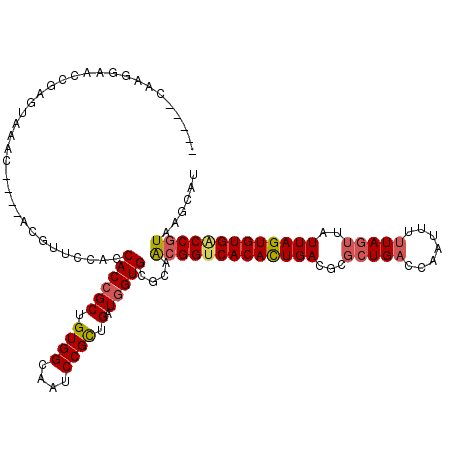
| Location | 2,442,076 – 2,442,196 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.25 |
| Mean single sequence MFE | -37.75 |
| Consensus MFE | -32.00 |
| Energy contribution | -33.50 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.896831 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2442076 120 + 22224390 CUGUGGCAAUCCGUGGCUGGUGCGCAACGGCCACACUGACGCGCUGACCAAUUUUUAGUUAUUAGUGUGACCUUAAGCAUUAUUAUUUUCACGUGUGGCACGCAAACGAGCCAAAAAACC .............(((((.((..((....((((((((((((((((((..(((.....))).))))))))....(((......)))...))).)))))))..))..)).)))))....... ( -34.60) >DroSec_CAF1 2973 120 + 1 CUGUGGCAAUCCGCUGAUGGUGCGCAGCGGUCACAUUGACGCGCUGACCAAUUUUUAGUUAUUAGUGUGACCGUAUGCAUUAUUAUUUUCACGUGUGGCACGCAAACGAGCCAAAAAACC ..((((....))))((((((((((..(((((((((((((...(((((.......)))))..))))))))))))).)))))))))).......((.((((.((....)).))))....)). ( -41.20) >DroSim_CAF1 5126 120 + 1 CUGUGGCAAUCCGCUGAUGGUGCGCAACGGUCACAUUGACGCGCUGACCAAUUUUUAGUUAUUAGUGUGACCGUUAGCAUUAUUAUUUUCACGUGUGGCACGCAAACGAGCCAAAAAACC ..((((....))))(((((((((..((((((((((((((...(((((.......)))))..)))))))))))))).))))))))).......((.((((.((....)).))))....)). ( -42.60) >DroYak_CAF1 3501 118 + 1 CUGUGGCAAUCCGUUGCUUGUGCGCAACGGUCACACUGACG--CUGGGCACUUUGUAUUUAUUAGUGUGGCCGUAAUCAUUAUUAUUUUCACGUGUGGCACGCAAACGAGUCAAAAAACC ...((((....((((((..((((...(((((((((((((..--....((.....)).....)))))))))))))...............((....)))))))).)))).))))....... ( -32.60) >consensus CUGUGGCAAUCCGCUGAUGGUGCGCAACGGUCACACUGACGCGCUGACCAAUUUUUAGUUAUUAGUGUGACCGUAAGCAUUAUUAUUUUCACGUGUGGCACGCAAACGAGCCAAAAAACC ..(((((.....))(((((((((...(((((((((((((...(((((.......)))))..)))))))))))))..)))))))))....)))((.((((.((....)).))))....)). (-32.00 = -33.50 + 1.50)

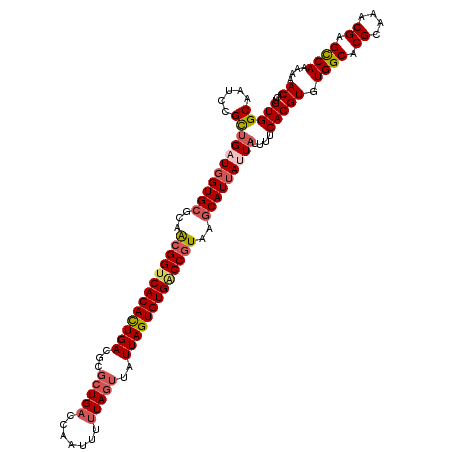
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:55:29 2006