
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 21,305,585 – 21,305,684 |
| Length | 99 |
| Max. P | 0.591470 |

| Location | 21,305,585 – 21,305,684 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.65 |
| Mean single sequence MFE | -25.30 |
| Consensus MFE | -14.49 |
| Energy contribution | -15.43 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.591470 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21305585 99 - 22224390 GCGAAAAUUACUUUAUAACAGCCA----AG-----UUGGCUGACUGAA-ACGCAGCGGCUGGGA-GUGGGAAAAUCCCUAAGGGAAUGGAAAGGCUGCAAAACAACUGAA---------- ..................(((((.----..-----(..((((.(((..-...)))))))..)..-..(((.....)))..............))))).............---------- ( -23.80) >DroSec_CAF1 47097 96 - 1 GCGAAAAUUACUUUAUAACAGCCA----AG-----UUGGCUGACUGAA-ACGCAGCGGCUGAGA-GCUGCAAAAUCCCUGGGGGAAUGG---GGCUGCAAAACAACUGAA---------- (((...............(((((.----..-----..)))))......-..((.(((((.....-)))))....((((...))))....---.)))))............---------- ( -25.20) >DroSim_CAF1 43526 96 - 1 GCGAAAAUUACUUUAUAACAGCCA----AG-----UUGGCUGACUGAA-ACGCAGCGGCUGAGA-GCGGGAAAAUCCCUGGGGGAAUGG---GGCUGNNNNNNNNNNNNN---------- ..................(((((.----..-----(..((((.(((..-...)))))))..)..-..(((.....)))...........---))))).............---------- ( -23.50) >DroEre_CAF1 45039 96 - 1 GCGAAAAUUACUUUAUAACAGCCA----AG-----UUGGUUGACUGACUCAGCAGCGGCUGAGAUGCGGGAAAAGUCCUG--GAACUGG---GGCUGCAAAACAACUGAA---------- ((................(((((.----..-----..))))).(((.((((((....))))))...)))....((((((.--.....))---))))))............---------- ( -27.00) >DroYak_CAF1 48375 98 - 1 GUGAAAAUUACUUUAUAACAGCCAACCAAG-----UUGGCUGACUGAA-AAGCAGCGGCGGAGA-GCGGGAAAAGUCCUG--GAAAUGG---GGCUGCAAAACAACUGAA---------- ..................((((((((...)-----)))))))......-..(((((..((....-.(((((....)))))--....)).---.)))))............---------- ( -26.90) >DroAna_CAF1 38857 108 - 1 UCGAAAAUUACUUUAUAACAGCCA----AGUAAAUUUGGCUGACUAAC-AAAGUACAGAUGUGG-CGGGUAAAACGCGUU---GAAAAG---GGGUGCAACAGGGUGUACAGAUGGGGGG ..........((((((..((((((----((....))))))))......-...(((((......(-((.......)))(((---(.....---.....))))....)))))..)))))).. ( -25.40) >consensus GCGAAAAUUACUUUAUAACAGCCA____AG_____UUGGCUGACUGAA_AAGCAGCGGCUGAGA_GCGGGAAAAUCCCUG__GGAAUGG___GGCUGCAAAACAACUGAA__________ ..................(((((............(((((((.(((......))))))))))....((((......))))............)))))....................... (-14.49 = -15.43 + 0.95)

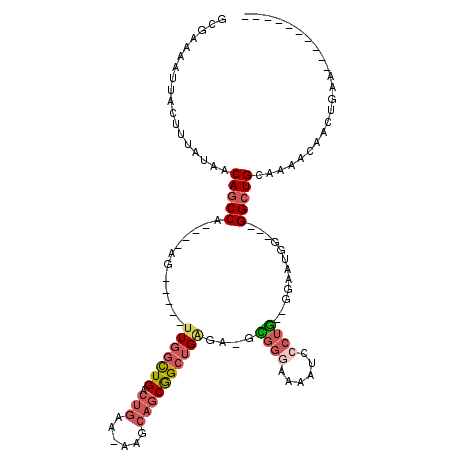

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:07:54 2006