| Sequence ID | X_DroMel_CAF1 |
|---|---|
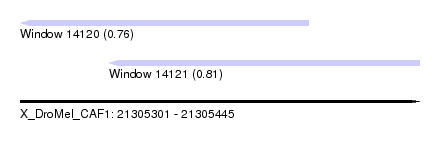
| Location | 21,305,301 – 21,305,445 |
| Length | 144 |
| Max. P | 0.812008 |

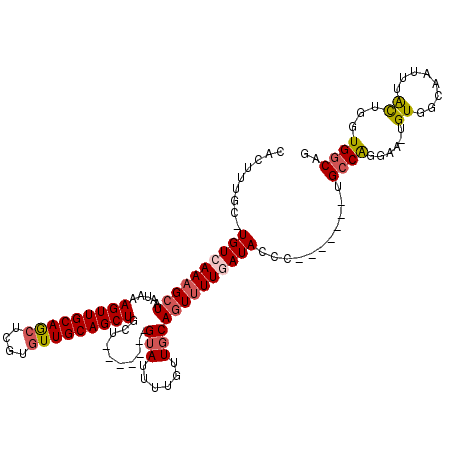
| Location | 21,305,301 – 21,305,405 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.97 |
| Mean single sequence MFE | -33.40 |
| Consensus MFE | -20.94 |
| Energy contribution | -21.54 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.757197 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21305301 104 - 22224390 CACUUUGC-UGUCAAAGCUAAUAAAGUUGCAGCUCGUAUUGCAGCUACU-------GUAUUUUGUUGCAGUUUUCAUACCC-------UGGCGGGAA-UGUGGCAACUAAUUGGUGGCAU .....(((-..((((.........((((((((......))))))))...-------.......(((((......(((.(((-------....))).)-))..)))))...))))..))). ( -29.80) >DroSec_CAF1 46814 105 - 1 CACUUUGCUUGUCAAAGCUAAUAAAGUUGCAGCUCGUGUUGCAGCUACU-------GUAUUUUGUUGCAGUUUUCAUACCC-------UGCCGGGAA-UGUGGCAACUAAUUGGUGGCAU .(((((((((....))))....)))))((((((....))))))((((((-------(.(((..(((((......(((.(((-------....))).)-))..))))).)))))))))).. ( -32.20) >DroEre_CAF1 44765 104 - 1 CACUUUGC-UGUCAAAGCUAAUAAAGUUGCAGCUCGUGUUGCAGCUGCA-------GUAUUUUGUUGCAGUUUUGAUACCC-------UGCCAGGAA-UGUUGCAAUUUACUGAUGGCAG ........-((((((((((.....(((((((((....)))))))))(((-------(.......))))))))))))))..(-------(((((.(((-(......)))).....)))))) ( -34.40) >DroYak_CAF1 48093 104 - 1 CACUUUGC-UGUCAAAGCUAAUAAAGUUGCAGCUCGUGUUGCAGCUGCU-------GUAUUUUGUUGCAGUUUUGAUACCC-------UGCCAGGAA-UGUUGCAAUCUACUGAUGGCAG .....(((-(((((.(((......(((((((((....))))))))))))-------(((..((((.(((.(((((......-------...))))).-))).))))..))))))))))). ( -32.50) >DroAna_CAF1 38591 118 - 1 CACUUGGC-UGUCAAAGCUAAUAAAGUUGCAACUCGUGUUGCAGCUGCUUUUUGCCGGAUUUUGUUGCAAUGUUGAUACCCCAAGCCCCGCCACAAAUUGUGGCAAU-UGCUAGUGGCAG ((((((((-(.....))))))...(((((((((....)))))))))((...((((((.(.(((((.((...(((.........)))...)).))))).).)))))).-.))..))).... ( -38.10) >consensus CACUUUGC_UGUCAAAGCUAAUAAAGUUGCAGCUCGUGUUGCAGCUGCU_______GUAUUUUGUUGCAGUUUUGAUACCC_______UGCCAGGAA_UGUGGCAAUUUACUGGUGGCAG .........((((((((((.....(((((((((....)))))))))..........(((......)))))))))))))...........((((......((........))...)))).. (-20.94 = -21.54 + 0.60)


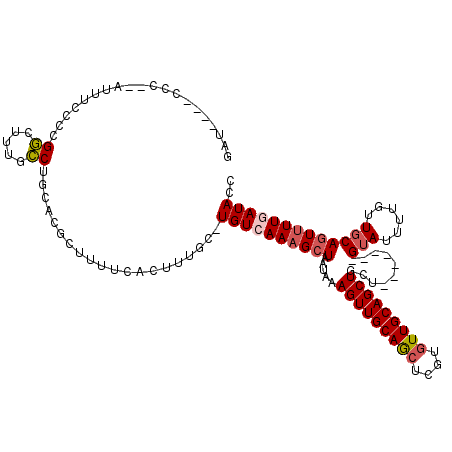
| Location | 21,305,333 – 21,305,445 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.26 |
| Mean single sequence MFE | -26.22 |
| Consensus MFE | -19.16 |
| Energy contribution | -19.88 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.812008 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21305333 112 - 22224390 GAUCUCACCCUGAUUUCCUCGGCUUUGCCUGCACGCUUUUCACUUUGC-UGUCAAAGCUAAUAAAGUUGCAGCUCGUAUUGCAGCUACU-------GUAUUUUGUUGCAGUUUUCAUACC ((((.......)))).....(((((((.(.(((............)))-.).))))))).....((((((((......))))))))(((-------(((......))))))......... ( -24.70) >DroSec_CAF1 46846 111 - 1 GAU--CACCCCGAUUUCCCCGGCAUUGCCUGCACGCUUUUCACUUUGCUUGUCAAAGCUAAUAAAGUUGCAGCUCGUGUUGCAGCUACU-------GUAUUUUGUUGCAGUUUUCAUACC (((--(.....)))).....(((.(((...(((.((..........)).)))))).))).....(((((((((....)))))))))(((-------(((......))))))......... ( -25.10) >DroEre_CAF1 44797 98 - 1 --------------UACCCCGGAUUUUCCUGCACGCUUUUCACUUUGC-UGUCAAAGCUAAUAAAGUUGCAGCUCGUGUUGCAGCUGCA-------GUAUUUUGUUGCAGUUUUGAUACC --------------......((.....)).....((..........))-((((((((((.....(((((((((....)))))))))(((-------(.......)))))))))))))).. ( -26.70) >DroYak_CAF1 48125 110 - 1 GAU--UUCCCCCAUUUCCCCGGCUUUUCCAGCACGCUUCUCACUUUGC-UGUCAAAGCUAAUAAAGUUGCAGCUCGUGUUGCAGCUGCU-------GUAUUUUGUUGCAGUUUUGAUACC ...--...............((((((..(((((............)))-))..)))))).....(((((((((....)))))))))(((-------(((......))))))......... ( -30.70) >DroAna_CAF1 38630 102 - 1 -----------------CCCGCCUCACGCCACCCACUUUUCACUUGGC-UGUCAAAGCUAAUAAAGUUGCAACUCGUGUUGCAGCUGCUUUUUGCCGGAUUUUGUUGCAAUGUUGAUACC -----------------..........((((.............))))-((((((.((.((((((((((((((....)))))))))((.....))......)))))))....)))))).. ( -23.92) >consensus GAU____CCC__AUUUCCCCGGCUUUGCCUGCACGCUUUUCACUUUGC_UGUCAAAGCUAAUAAAGUUGCAGCUCGUGUUGCAGCUGCU_______GUAUUUUGUUGCAGUUUUGAUACC ....................((.....))....................((((((((((.....(((((((((....)))))))))..........(((......))))))))))))).. (-19.16 = -19.88 + 0.72)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:07:53 2006