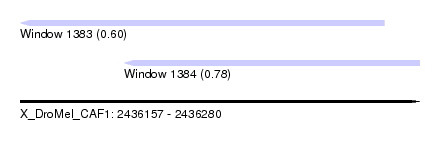
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,436,157 – 2,436,280 |
| Length | 123 |
| Max. P | 0.777191 |

| Location | 2,436,157 – 2,436,269 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 82.86 |
| Mean single sequence MFE | -38.63 |
| Consensus MFE | -26.96 |
| Energy contribution | -27.88 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.603553 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
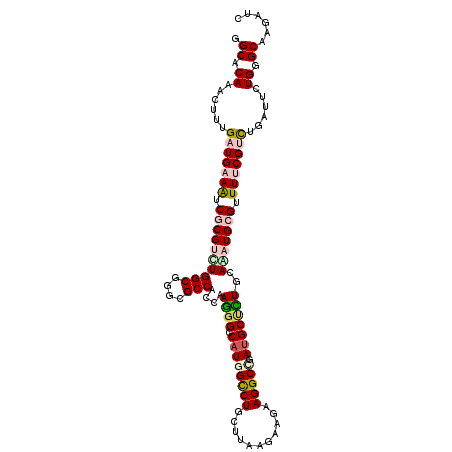
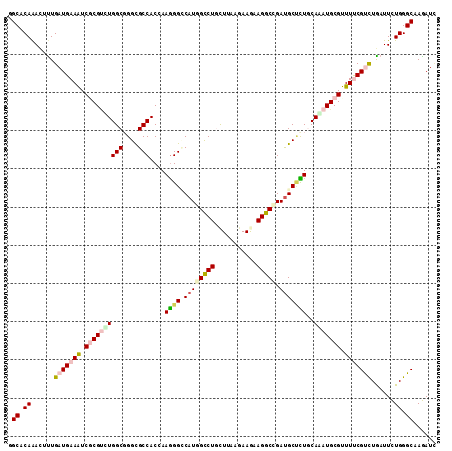
>X_DroMel_CAF1 2436157 112 - 22224390 GGCCCAAACGCUGAUGAAGUCGCGUUUGGCGGGCGCCACCAAGGGACAUGGUCUGCUAAAGAAAAAGGCGGAUGCCCUGCAGAUGCGCUUUCGUCUGAUUCUGGGCAAGAUC .(((((....(.(((((((.((((((((..(((((((......)).)...(((((((.........)))))))))))..)))))))).))))))).)....)))))...... ( -45.90) >DroVir_CAF1 171054 112 - 1 GGCACAAACUUUGAUGAAAUCGCGUCUGGCGGGCGCCACAAAGGGCCAUGGCCUGCUUAAGAAGAAGGCCGAUGCGCUGCAGAUGCGUUUUCGCCUGAUUCUGGGCAAGAUC ...............((((.((((((((((((((.((.....))))).((((((.((.....)).))))))...)))..)))))))).))))((((......))))...... ( -43.00) >DroGri_CAF1 126453 112 - 1 CGCACAAAUUUUGAUGAAAUCGCGCCUGGCGGGCGCCACCAAGGGUCAUGGCCUGCUUAAAAAGAAGGCAGAUGCUCUCCAAAUGCGUUUUCGCCUAAUUCUGGGCAAGAUC ...............((((.((((..(((.((((((((((...)))...)))((((((.......))))))..)))).)))..)))).))))(((((....)))))...... ( -36.10) >DroWil_CAF1 111701 112 - 1 GGCCCAAACUUUGAUGAAAUCUCGAUUGGCUGGUGCCACCAAGGGCCAUGGUCUGUUAAAAAAGAAGGCCGAUGCCUUGCAAAUGCGUUUCCGUUUGAUUUUGGGCAAAAUU (((((.....((((.(....))))).((((....))))....))))).((((((...........)))))).(((((..((((((......)))))).....)))))..... ( -35.00) >DroMoj_CAF1 164605 112 - 1 AGCACAAACGUUGAUGAAAUCGCGUCUGGCGGGCGCCACGAAAGGCCAUGGCCUACUUAAGAAGAAGGCUGACGCUUUGCAGAUGCGUUUCCGUUUGAUUCUGGGCAAGAUC ....((((((.....((((.((((((((..(((((((......)))...(((((.((.....)).)))))...))))..))))))))))))))))))..(((.....))).. ( -36.40) >DroAna_CAF1 94135 112 - 1 AGCUCAGACAUUGAUGAAAUCCCGUCUGGCUGGUGCCACAAAGGGACAUGGCCUCCUCAAGAAAAAGGCCGAUGCUCUACAAAUGCGUUUUCGUUUAAUUUUGGGCAAAAUA .(((((((....((((((.((((...((((....))))....))))...(((((...........)))))(((((.........)))))))))))....)))))))...... ( -35.40) >consensus GGCACAAACUUUGAUGAAAUCGCGUCUGGCGGGCGCCACCAAGGGCCAUGGCCUGCUUAAGAAGAAGGCCGAUGCUCUGCAAAUGCGUUUUCGUCUGAUUCUGGGCAAGAUC .((.((......(((((((.((((((((((....)))....((((.((((((((...........))))).)))))))..))))))).)))))))......)).))...... (-26.96 = -27.88 + 0.92)

| Location | 2,436,189 – 2,436,280 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 78.05 |
| Mean single sequence MFE | -30.20 |
| Consensus MFE | -20.38 |
| Energy contribution | -20.68 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.777191 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
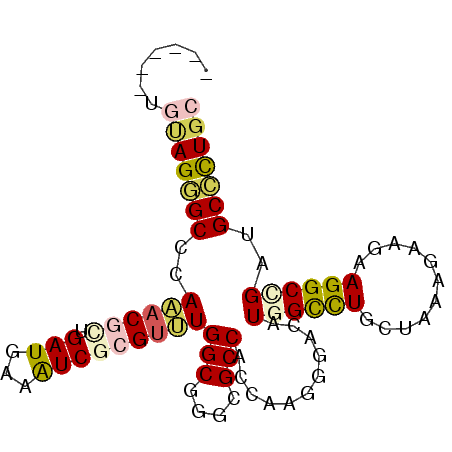
>X_DroMel_CAF1 2436189 91 - 22224390 CCAUGUUGUAGGGCCCAAACGCUGAUGAAGUCGCGUUUGGCGGGCGCCACCAAGGGACAUGGUCUGCUAAAGAAAAAGGCGGAUGCCCUGC .......(((((((((((((((.(((...)))))))))))....((((.(((.......)))(((.....)))....))))...))))))) ( -33.50) >DroEre_CAF1 84079 84 - 1 -------GUAGUGCCCAAACGCUGAUGAAGUCGCGUUUGGCGGGCGCCACCAAGGGGCAUGGCCUGCUGAAGAAGAAGGCAGACGCCCUGC -------((.((((((((((((.(((...)))))))))...)))))).))...(((((...((((.((.....)).))))....))))).. ( -34.70) >DroWil_CAF1 111733 87 - 1 CA----UUUAGGGCCCAAACUUUGAUGAAAUCUCGAUUGGCUGGUGCCACCAAGGGCCAUGGUCUGUUAAAAAAGAAGGCCGAUGCCUUGC ..----.....(((((.....((((.(....))))).((((....))))....))))).((((((...........))))))......... ( -24.10) >DroYak_CAF1 79321 89 - 1 CC--AUUGUAGGGCACAAACGCUGAUGAAGUCGCGUUUGGCCGGCGCCACCAAGGGACAUGGUUUGCUCAAGAAGAAGGCCGAUGCCCUGC ..--...(((((((((((((((.(((...))))))))))..((((((.((((.......))))..))((.....))..)))).)))))))) ( -33.50) >DroMoj_CAF1 164637 85 - 1 ------CGCAGAGCACAAACGUUGAUGAAAUCGCGUCUGGCGGGCGCCACGAAAGGCCAUGGCCUACUUAAGAAGAAGGCUGACGCUUUGC ------.(((((((.....((((((((......)))).))))(((....(....))))..(((((.((.....)).)))))...))))))) ( -27.50) >DroAna_CAF1 94167 82 - 1 ---------AGAGCUCAGACAUUGAUGAAAUCCCGUCUGGCUGGUGCCACAAAGGGACAUGGCCUCCUCAAGAAAAAGGCCGAUGCUCUAC ---------(((((((..............((((...((((....))))....))))...(((((...........))))))).))))).. ( -27.90) >consensus ______UGUAGGGCCCAAACGCUGAUGAAAUCGCGUUUGGCGGGCGCCACCAAGGGACAUGGCCUGCUAAAGAAGAAGGCCGAUGCCCUGC .......(((((((..((((((.(((...)))))))))(((....)))...........((((((...........))))))..))))))) (-20.38 = -20.68 + 0.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:55:27 2006