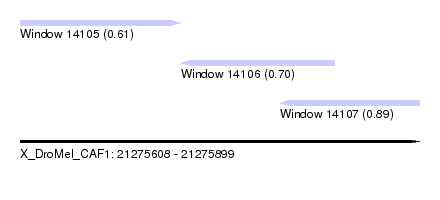
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 21,275,608 – 21,275,899 |
| Length | 291 |
| Max. P | 0.886650 |

| Location | 21,275,608 – 21,275,725 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.21 |
| Mean single sequence MFE | -35.04 |
| Consensus MFE | -16.53 |
| Energy contribution | -17.32 |
| Covariance contribution | 0.79 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.606509 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
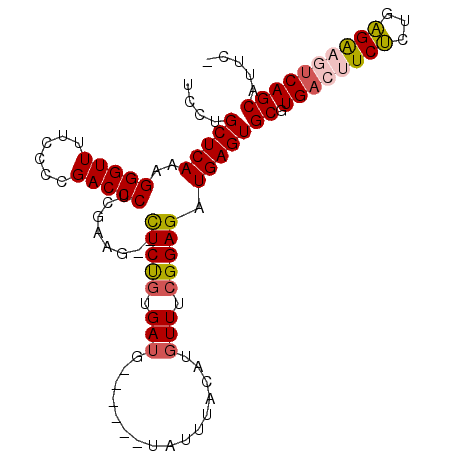
>X_DroMel_CAF1 21275608 117 + 22224390 UUACAUUAGCGGUGGGUGCGUGAGUGGAUUUUAAUUUUCAAUAAGAAGACUUCAAAAUCACUCUAU--UUGCGCUGCAAGCCCGACUUGGGCUCAG-UACUCAAGCUGAGCAAAUUGCUU .....(((((..((((((((.((((((.(((.(((((((.....))))).)).))).))))))...--.)))((((..(((((.....))))))))-)))))).)))))((.....)).. ( -35.30) >DroSec_CAF1 16753 117 + 1 UCACAUCAGCGGUGGGUGCGUGAGUGGAUUUUAAUUUUCAAUAAGAAGACUUCAAAAUCACUUAAU--UUGCGCUGCAAGCCCGACUUGUGCUCAA-UACUCAAGCUGAGCAAGUUGCUU .....(((((..((((((((..(((.(((((((((((((.....))))).)).)))))).....))--)..))).(((((.....)))))......-.))))).)))))((.....)).. ( -31.00) >DroSim_CAF1 28273 117 + 1 UUACAUCAGCGGUGGGUGCGUGAGUGGAUUUUAAUUUUCAAUAAGAAGACUUCAAAAUCACUUAAU--UUGCGCUGCAAGCCCGGCUUGGGCUCAG-UACUCAAGCUGAGCAAGUUGCUU .....(((((..(((((((.(((((((.(((.(((((((.....))))).)).))).)))))))..--.((.(((.(((((...)))))))).)))-)))))).)))))((.....)).. ( -36.10) >DroEre_CAF1 16772 102 + 1 UUACAUCAGCGGUGGGAGCGUGAGUGGAGUC----------------GGCUUCCGAAUCACUCCGC--UCACGGUGCAAGCCCGACUUGGGCGCAGCUACUCCGGCUAAGCAAAUUGAUU ........(((((.(((((((((((((((((----------------((...))))....))))))--)))))..((..((((.....))))...))..)))).)))..))......... ( -43.30) >DroYak_CAF1 18629 102 + 1 UUACAUCAGCGGUGGGAGCGUGAGUGGAUUUUAAUUUCCAAUAAACGGGCUUCGAAAGCACUCCACUCUCACGAUGCUAGCCCGACUUGGGAACA------------AAGCAGA------ ...........(((((((.(.(((((..(((..(..(((.......)))..)..))).)))))).)))))))..((((..(((.....)))....------------.))))..------ ( -29.50) >consensus UUACAUCAGCGGUGGGUGCGUGAGUGGAUUUUAAUUUUCAAUAAGAAGACUUCAAAAUCACUCAAU__UUGCGCUGCAAGCCCGACUUGGGCUCAG_UACUCAAGCUGAGCAAAUUGCUU ........((((((.(...(((((((.(((((..(((((.....)))))....)))))))))).))...).))))))..((((.....))))............................ (-16.53 = -17.32 + 0.79)

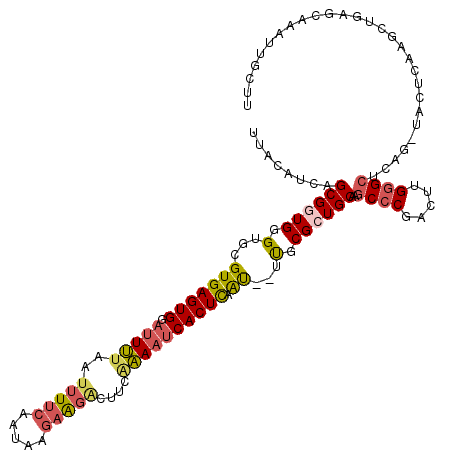
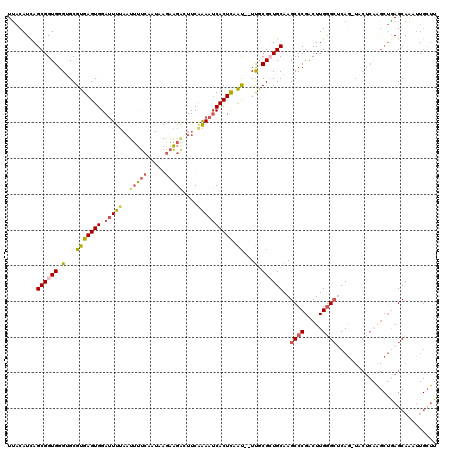
| Location | 21,275,725 – 21,275,837 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 81.10 |
| Mean single sequence MFE | -28.64 |
| Consensus MFE | -21.39 |
| Energy contribution | -21.50 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.696643 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21275725 112 - 22224390 CGGAGAUGAGUGCGUGACUUCUCUGAGAAGUCAGCAUUCAUGCUAACCAACUCUCUUUCUCUUUCUCCAGAAAUAUAUUU-UUUCGCAGGGUCCCGUCUCUGUUUUGUUUCGG .((((((((((((.(((((((.....))))))))))))))((.....))..............))))).((((((.....-....((((((......))))))..)))))).. ( -33.20) >DroSec_CAF1 16870 102 - 1 CGGAGAUGAGUGCGUGACUUCUCUGAGAAGUCAGCAUUCACGCUAAUCAACUCUCU---------UCCAGGAAUAUAUUUCUUUCGUAGGGCCCAGU--CUGUUUUGUUUCGG (((((.(((((((.(((((((.....))))))))))))))....(((..(((((((---------...(((((.....)))))....))))...)))--..)))...))))). ( -26.30) >DroSim_CAF1 28390 88 - 1 CGGAGAUGAGUGCGUGACUUCUCUGAGAAGUCAGCAUU--------------CUCU---------UCCAGGAAUAUAUUUCUUUCGUAGGGCCCAGU--CUGUUUUGUUUCGG .((((..((((((.(((((((.....))))))))))))--------------)..)---------))).((((((.............((((...))--))....)))))).. ( -26.43) >consensus CGGAGAUGAGUGCGUGACUUCUCUGAGAAGUCAGCAUUCA_GCUAA_CAACUCUCU_________UCCAGGAAUAUAUUUCUUUCGUAGGGCCCAGU__CUGUUUUGUUUCGG ..(((((((((((.(((((((.....)))))))))))))..........................(((.((((........))))...)))...............))))).. (-21.39 = -21.50 + 0.11)


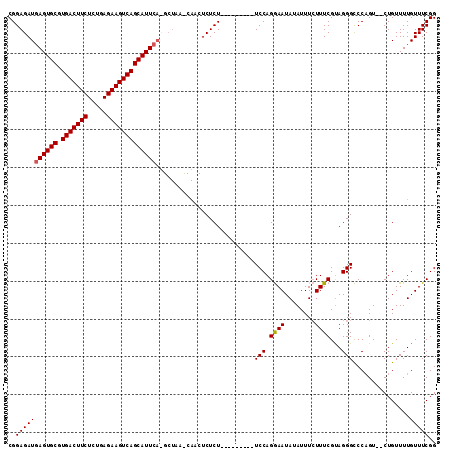
| Location | 21,275,797 – 21,275,899 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 87.75 |
| Mean single sequence MFE | -31.28 |
| Consensus MFE | -23.05 |
| Energy contribution | -23.49 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.886650 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21275797 102 - 22224390 UCCUGCUCAAAGGGUUUUCCCCGACCCGCAAAG--CUCUGUGAUGUACAUGUAUUUACAUUUUUCGGAGAUGAGUGCGUGACUUCUCUGAGAAGUCAGCAUUCA ...........(((((......)))))......--(((((.((((((........))))))...))))).(((((((.(((((((.....)))))))))))))) ( -34.90) >DroSec_CAF1 16932 96 - 1 UCCUGCUCAAAGGGUUUUCCCCGACCCUCGAAG--CUCUGUGAUG------UAUUUACAUGUUUCGGAGAUGAGUGCGUGACUUCUCUGAGAAGUCAGCAUUCA .....(((..((((((......))))))(((((--(..(((((..------...))))).))))))))).(((((((.(((((((.....)))))))))))))) ( -37.80) >DroSim_CAF1 28440 94 - 1 UCCUGCUCAAAGGGUUUUCCCCGACCCUCGAAG--CUCUGUGAUG------UAUUUACAUGUUUCGGAGAUGAGUGCGUGACUUCUCUGAGAAGUCAGCAUU-- ....(((((.((((((......))))))(((((--(..(((((..------...))))).))))))....)))))((.(((((((.....)))))))))...-- ( -33.70) >DroEre_CAF1 16921 96 - 1 UCCUGCUCAAAGGGUUUUCCCCGACCCCCGACGUAUUCCUUGAUG------UAUUUACAUGUUUCGGAGAUGAGUGCGUGACUUCCUUGGGG-CCCAGCACCC- ...((((....(((....((((((......(((((((((((((..------(((....)))..)).)))..)))))))).......))))))-)))))))...- ( -23.12) >DroYak_CAF1 18769 94 - 1 UCCUGCUCAAAGGGUUUUCCCCGACCCUCGCAG--UUCUGUGAUG------UAUUUACAUGUUUCGGAGAUGAGUGCGUGACUUCUCUGAGG-CUCAGCAUUC- ..((((....((((((......)))))).))))--...(((((..------...))))).((((((((((..(((.....))))))))))))-).........- ( -26.90) >consensus UCCUGCUCAAAGGGUUUUCCCCGACCCUCGAAG__CUCUGUGAUG______UAUUUACAUGUUUCGGAGAUGAGUGCGUGACUUCUCUGAGAAGUCAGCAUUC_ ....(((((..(((((......)))))........(((((.(((................))).))))).)))))((.((((((((...))))))))))..... (-23.05 = -23.49 + 0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:07:38 2006