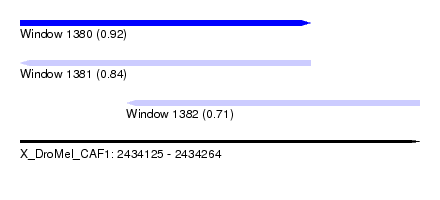
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,434,125 – 2,434,264 |
| Length | 139 |
| Max. P | 0.915259 |

| Location | 2,434,125 – 2,434,226 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 76.19 |
| Mean single sequence MFE | -20.23 |
| Consensus MFE | -11.50 |
| Energy contribution | -11.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915259 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2434125 101 + 22224390 --AGUUCGAUAAGUCGGAUAUCGAUCGAUACACGACACACACACACGCAUACAUACAUAUAUCCUAUAUUUUUGUAUAUUUACCAACAG----CAGUUGGGCCACAG --..........((((..((((....))))..))))..........(((((((...(((((...)))))...))))).....(((((..----..)))))))..... ( -21.10) >DroSec_CAF1 76869 92 + 1 --AGUUCGAUAAGUCGAAUAUCGAUCGAUACACGACACACACACACAUAUACAUA---------UAUAUUUUUGUAUAUUUACCAACAG----CAGUUGGGCCACAG --..........((((..((((....))))..))))..........(((((((..---------........)))))))...(((((..----..)))))....... ( -18.00) >DroEre_CAF1 81936 97 + 1 UAGGUUCGAUAAAUCG-AUAUCGAUCGACAUG---CAUACACACAC------AUAUGUAUAUCCUAUAUUUUUGUAUAUUCACCAACAACAAACAGUUGGGUCACCG ..(((((((((.....-.))))))..((((((---((((.......------.)))))))....(((((....)))))....(((((........))))))))))). ( -21.60) >consensus __AGUUCGAUAAGUCG_AUAUCGAUCGAUACACGACACACACACAC__AUACAUA__UAUAUCCUAUAUUUUUGUAUAUUUACCAACAG____CAGUUGGGCCACAG ...(.((((((.......)))))).)......................................(((((....)))))....(((((........)))))....... (-11.50 = -11.50 + 0.00)

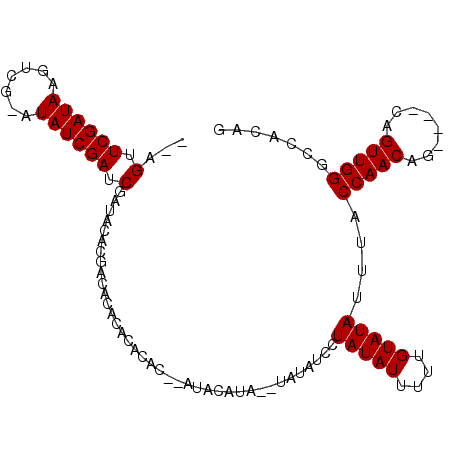
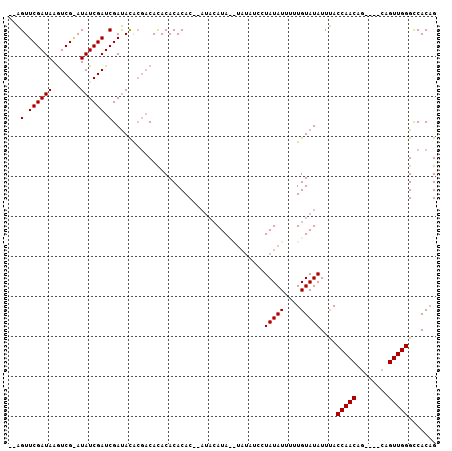
| Location | 2,434,125 – 2,434,226 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 76.19 |
| Mean single sequence MFE | -24.87 |
| Consensus MFE | -15.66 |
| Energy contribution | -17.33 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.836415 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2434125 101 - 22224390 CUGUGGCCCAACUG----CUGUUGGUAAAUAUACAAAAAUAUAGGAUAUAUGUAUGUAUGCGUGUGUGUGUGUCGUGUAUCGAUCGAUAUCCGACUUAUCGAACU-- .((((..(((((..----..)))))......))))...(((((.((((((..((..(....)..))..)))))).)))))...((((((.......))))))...-- ( -24.90) >DroSec_CAF1 76869 92 - 1 CUGUGGCCCAACUG----CUGUUGGUAAAUAUACAAAAAUAUA---------UAUGUAUAUGUGUGUGUGUGUCGUGUAUCGAUCGAUAUUCGACUUAUCGAACU-- .......(((((..----..)))))...(((((((...(((((---------(....)))))).)))))))((((.(((((....))))).))))..........-- ( -24.00) >DroEre_CAF1 81936 97 - 1 CGGUGACCCAACUGUUUGUUGUUGGUGAAUAUACAAAAAUAUAGGAUAUACAUAU------GUGUGUGUAUG---CAUGUCGAUCGAUAU-CGAUUUAUCGAACCUA .(((((((((((........)))))((.(((((((...((((((......).)))------)).))))))).---)).)))..((((((.-.....))))))))).. ( -25.70) >consensus CUGUGGCCCAACUG____CUGUUGGUAAAUAUACAAAAAUAUAGGAUAUA__UAUGUAU__GUGUGUGUGUGUCGUGUAUCGAUCGAUAU_CGACUUAUCGAACU__ .......(((((........)))))...(((((((...(((((...........)))))...)))))))..((((.(((((....))))).))))............ (-15.66 = -17.33 + 1.67)

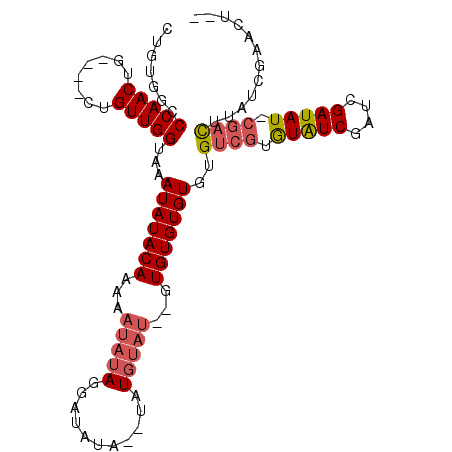
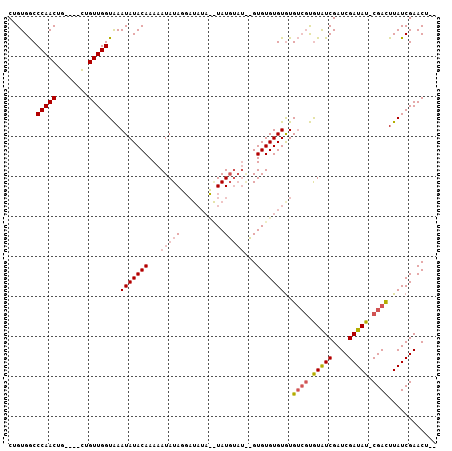
| Location | 2,434,162 – 2,434,264 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 81.13 |
| Mean single sequence MFE | -22.07 |
| Consensus MFE | -15.85 |
| Energy contribution | -15.97 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.708154 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
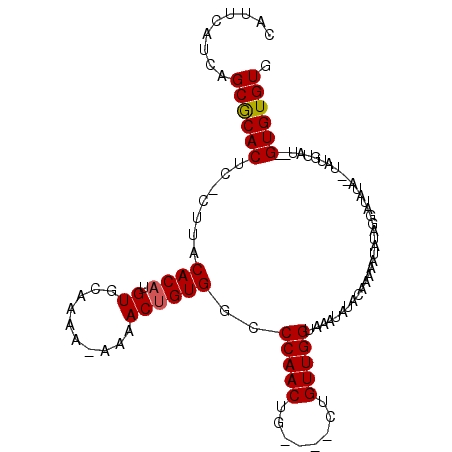
>X_DroMel_CAF1 2434162 102 - 22224390 CAUUCAUCAGCGCACUU-CUUACACAUGUGCAAAA-AAAACUGUGGCCCAACUG----CUGUUGGUAAAUAUACAAAAAUAUAGGAUAUAUGUAUGUAUGCGUGUGUG .........((((((..-....((((.((......-...))))))(((((((..----..))))).....(((((...(((((...)))))...))))))))))))). ( -22.30) >DroSec_CAF1 76906 94 - 1 CAUUCAUCAGCGCACUCCCUUACACAAGUGCAAAA-AAAACUGUGGCCCAACUG----CUGUUGGUAAAUAUACAAAAAUAUA---------UAUGUAUAUGUGUGUG .........((((((.......((((.((......-...))))))..(((((..----..)))))...(((((((........---------..))))))))))))). ( -20.30) >DroEre_CAF1 81971 101 - 1 CAUUCAUCAGCACACUC-CUUACACAUGUGCAAAAAAAAACGGUGACCCAACUGUUUGUUGUUGGUGAAUAUACAAAAAUAUAGGAUAUACAUAU------GUGUGUG .((((((((((((((..-.........))).......(((((((......)))))))..)))))))))))(((((...((((.(......)))))------.))))). ( -23.60) >consensus CAUUCAUCAGCGCACUC_CUUACACAUGUGCAAAA_AAAACUGUGGCCCAACUG____CUGUUGGUAAAUAUACAAAAAUAUAGGAUAUA__UAUGUAU__GUGUGUG .........((((((.......((((.((..........))))))..(((((........)))))....................................)))))). (-15.85 = -15.97 + 0.11)

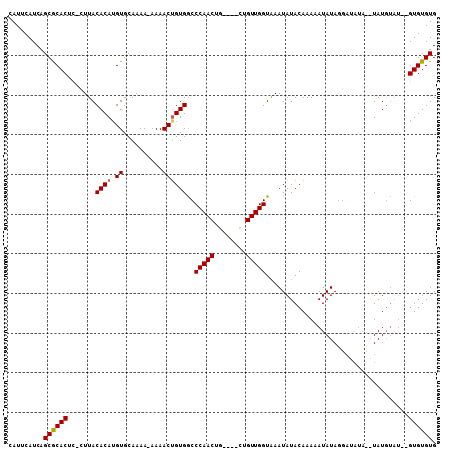
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:55:26 2006