
| Sequence ID | X_DroMel_CAF1 |
|---|---|
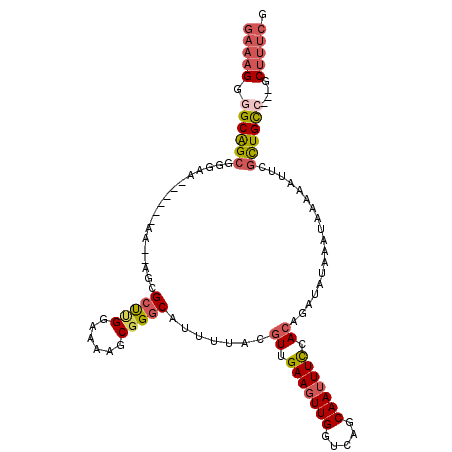
| Location | 21,229,344 – 21,229,445 |
| Length | 101 |
| Max. P | 0.987492 |

| Location | 21,229,344 – 21,229,445 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 79.46 |
| Mean single sequence MFE | -35.35 |
| Consensus MFE | -21.66 |
| Energy contribution | -22.38 |
| Covariance contribution | 0.73 |
| Combinations/Pair | 1.24 |
| Mean z-score | -3.78 |
| Structure conservation index | 0.61 |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.987492 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21229344 101 + 22224390 GAAAGGGGCAGCGGGAG------AA--AGCGCUUGGAAAAGCGGGCAUUUUACGUUGAAGUUGGUCAGCAAUUUCCACAGAUAUAAAUAAAAAUUCGCUGCC---GCUUUCG (((((.(((((((((..------..--..(((((....)))))..........((.(((((((.....))))))).))...............)))))))))---.))))). ( -39.20) >DroSec_CAF1 41801 103 + 1 GAAAGGAGCAGCUGGAA------AAGCAGAGCUCGGAAAAGCGGGCAUUUUACGUUGAAGUUGGUCAGCAAUUUCCACAGAUAUAAAUAAAAAUUCGCUGCC---GCUUUCG (((((..((((((((((------(......(((((......))))).......(((((......)))))..))))))..((.............))))))).---.))))). ( -31.12) >DroSim_CAF1 33251 103 + 1 GAAAGGGGCAGCGGGAA------AAGCAGCGCUCGGAAAAGCGGGCAUUUUACGUUGAAGUUGGUCAGCAAUUUCCACAGAUAUAAAUAAAAAUUCGCUGCC---GCUUUCG (((((.(((((((((..------.......(((((......))))).......((.(((((((.....))))))).))...............)))))))))---.))))). ( -39.00) >DroEre_CAF1 27572 99 + 1 GAAUGGGGCAGC--------GGAAA--AGCGCCUGGAAAAGCGGGCAUUUUAAGUUGAAGUUGGUCAGCAAUUUCUACAGAUAUAAAUAAAAAUUCGCUGCC---GCUUUCG (((.(.((((((--------(((..--...(((((......))))).......((.(((((((.....))))))).))...............)))))))))---.).))). ( -36.40) >DroYak_CAF1 28531 107 + 1 GAAAGGGGCGGCGGGAAAGCGGGAA--AGCGCCUGGAAAAGCGGGCAUUUAACGUUGAAGUUGGUCAGCAAUUUCAACAGAUAUAAAUAAAAAUUCGCUGUC---GCUUUCG (((((.(((((((((...((.....--.))(((((......))))).......((((((((((.....))))))))))...............)))))))))---.))))). ( -42.20) >DroAna_CAF1 26088 99 + 1 AACAGGAGCGGA----G------UGGUAGUGGUGGGGGUAGCAGGCUGGCUAAAUGGAAGUUGGUCAGCAAGUUUUACAGUUAUAAAUACAAAUUUGUUGUCUAGGCUU--- ...(((((((((----.------..(((...((((..(((((..(((((((((.......)))))))))..))..)))..))))...)))...)))))).)))......--- ( -24.20) >consensus GAAAGGGGCAGCGGGAA______AA__AGCGCUUGGAAAAGCGGGCAUUUUACGUUGAAGUUGGUCAGCAAUUUCCACAGAUAUAAAUAAAAAUUCGCUGCC___GCUUUCG (((((.((((((..................(((((......))))).......((.(((((((.....))))))).))..................))))))....))))). (-21.66 = -22.38 + 0.73)


| Location | 21,229,344 – 21,229,445 |
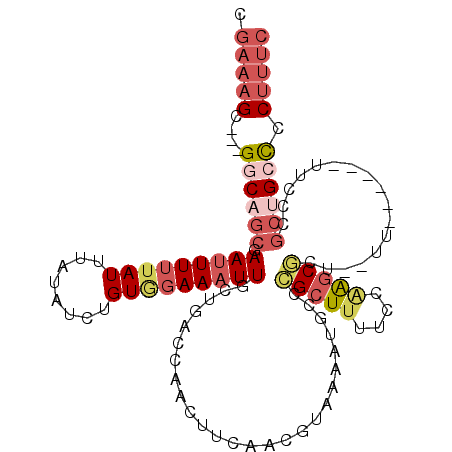
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 79.46 |
| Mean single sequence MFE | -23.03 |
| Consensus MFE | -12.22 |
| Energy contribution | -13.88 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.53 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.506980 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21229344 101 - 22224390 CGAAAGC---GGCAGCGAAUUUUUAUUUAUAUCUGUGGAAAUUGCUGACCAACUUCAACGUAAAAUGCCCGCUUUUCCAAGCGCU--UU------CUCCCGCUGCCCCUUUC .(((((.---((((((((((((((((........)))))))))(((((......))).............((((....)))))).--..------....))))))).))))) ( -27.50) >DroSec_CAF1 41801 103 - 1 CGAAAGC---GGCAGCGAAUUUUUAUUUAUAUCUGUGGAAAUUGCUGACCAACUUCAACGUAAAAUGCCCGCUUUUCCGAGCUCUGCUU------UUCCAGCUGCUCCUUUC .((((((---((((((.(((((((((........)))))))))))))....((......)).......))))))))..((((...(((.------....))).))))..... ( -27.10) >DroSim_CAF1 33251 103 - 1 CGAAAGC---GGCAGCGAAUUUUUAUUUAUAUCUGUGGAAAUUGCUGACCAACUUCAACGUAAAAUGCCCGCUUUUCCGAGCGCUGCUU------UUCCCGCUGCCCCUUUC .(((((.---((((((((((((((((........))))))))).......................((.(((((....)))))..))..------....))))))).))))) ( -29.50) >DroEre_CAF1 27572 99 - 1 CGAAAGC---GGCAGCGAAUUUUUAUUUAUAUCUGUAGAAAUUGCUGACCAACUUCAACUUAAAAUGCCCGCUUUUCCAGGCGCU--UUUCC--------GCUGCCCCAUUC .(((.(.---((((((((((((((((........)))))))))(((((......))).........(((.(......).))))).--....)--------)))))).).))) ( -24.20) >DroYak_CAF1 28531 107 - 1 CGAAAGC---GACAGCGAAUUUUUAUUUAUAUCUGUUGAAAUUGCUGACCAACUUCAACGUUAAAUGCCCGCUUUUCCAGGCGCU--UUCCCGCUUUCCCGCCGCCCCUUUC .((((((---(..(((.......((((((....(((((((.(((.....))).))))))).))))))...(((......))))))--....))))))).............. ( -22.80) >DroAna_CAF1 26088 99 - 1 ---AAGCCUAGACAACAAAUUUGUAUUUAUAACUGUAAAACUUGCUGACCAACUUCCAUUUAGCCAGCCUGCUACCCCCACCACUACCA------C----UCCGCUCCUGUU ---.(((..((((((.....))))..........(((...((.(((((...........))))).))..))).................------)----)..)))...... ( -7.10) >consensus CGAAAGC___GGCAGCGAAUUUUUAUUUAUAUCUGUGGAAAUUGCUGACCAACUUCAACGUAAAAUGCCCGCUUUUCCAAGCGCU__UU______UUCCCGCUGCCCCUUUC .(((((....((((((.(((((((((........)))))))))..........................(((((....))))).................)))))).))))) (-12.22 = -13.88 + 1.67)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:07:28 2006