| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 21,212,841 – 21,212,943 |
| Length | 102 |
| Max. P | 0.806340 |

| Location | 21,212,841 – 21,212,943 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 83.43 |
| Mean single sequence MFE | -36.93 |
| Consensus MFE | -24.94 |
| Energy contribution | -24.63 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.98 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.806340 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
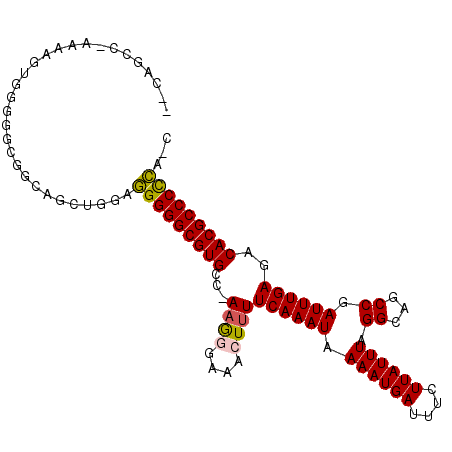
>X_DroMel_CAF1 21212841 102 + 22224390 --CUUCCAAAAAGUG-GGGUGGCAGCUGGAGGGGGCGUGCCAAAGGGAAACUUUUCAAAUAAAAUGAUUUCUUAUUUAGGCAACCGAUUUGAGACACGCCCCCA-C --.(((((.....))-)))...........(((((((((..(((((....)))))..............(((((.(..(....)..)..)))))))))))))).-. ( -32.50) >DroPse_CAF1 13341 98 + 1 CGCAGCC-CACAGGGGCGAGGGCA-----AAGGGGCGUGGA-AAACGAAACUUUUCAAAUAAAAUGAUUUCUUAUUUAGGCAUCCGAUUUGAGACACGCCCCUU-C .((.(((-(....))))....)).-----((((((((((((-(((......))))).............(((((....((...))....)))))))))))))))-. ( -35.20) >DroSim_CAF1 16657 101 + 1 --CCUCG-AAAAGUG-GGGCGGCAGCUGGAGGGGGCGUGCCAAAGGGAAACUUUUCAAAUAAAAUGAUUUCUUAUUUAGGCAGCCGAUUUGAGACACGCCCCCA-C --.....-....(.(-(((((....((.(((..(((.(((((((((....)))))......((((((....)))))).)))))))..))).))...))))))).-. ( -36.20) >DroEre_CAF1 11918 102 + 1 --CAGCG-AAGAGUGGGGGCGGCAGCUGGAGGGGGCGUGCCGAAGGGAAACUUUUCAAAUAAAAUGAUUUCUUAUUUAGGCAGCCGAUUUGAGCCACGCCCCCA-U --.....-....(((((((((...(((.(((..(((.(((((((((....)))))......((((((....)))))).)))))))..))).)))..))))))))-) ( -44.50) >DroYak_CAF1 12322 102 + 1 --CAGCG-AAAAGUGGGGGCGGCAGCUGGAGGGGGCGUGCCGAUGGGAAACUUUUCAAAUAAAAUGAUUUCUUAUUUAGGCAGCCGAUUUGAGACACGCCCCCA-U --.....-....(((((((((....((.(((..(((.((((((.((....))..)).....((((((....)))))).)))))))..))).))...))))))))-) ( -40.80) >DroPer_CAF1 13541 99 + 1 CGCAGCC-CACAGGGGCGAGGGCA-----AAGGGGCGUGGA-AAACGAAACUUUUCAAAUAAAAUGAUUUCUUAUUUAGGCAUCCGAUUUGAGACACGCCCUUCCC ....(((-(....))))..(((..-----.(((((((((((-(((......))))).............(((((....((...))....))))))))))))))))) ( -32.40) >consensus __CAGCC_AAAAGUGGGGGCGGCAGCUGGAGGGGGCGUGCC_AAGGGAAACUUUUCAAAUAAAAUGAUUUCUUAUUUAGGCAGCCGAUUUGAGACACGCCCCCA_C ..............................(((((((((...((((....))))((((((.((((((....)))))).((...)).))))))..)))))))))... (-24.94 = -24.63 + -0.30)


| Location | 21,212,841 – 21,212,943 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 83.43 |
| Mean single sequence MFE | -32.33 |
| Consensus MFE | -22.06 |
| Energy contribution | -22.04 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.702757 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
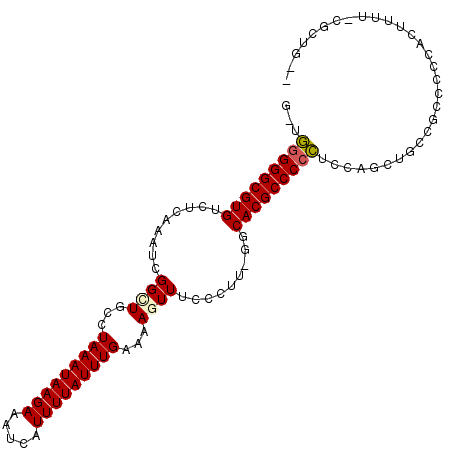
>X_DroMel_CAF1 21212841 102 - 22224390 G-UGGGGGCGUGUCUCAAAUCGGUUGCCUAAAUAAGAAAUCAUUUUAUUUGAAAAGUUUCCCUUUGGCACGCCCCCUCCAGCUGCCACCC-CACUUUUUGGAAG-- .-.(((((((((((.......((..((.(((((((((.....)))))))))....))..))....)))))))))))(((((.........-......)))))..-- ( -31.36) >DroPse_CAF1 13341 98 - 1 G-AAGGGGCGUGUCUCAAAUCGGAUGCCUAAAUAAGAAAUCAUUUUAUUUGAAAAGUUUCGUUU-UCCACGCCCCUU-----UGCCCUCGCCCCUGUG-GGCUGCG (-((((((((((....((((..(((...(((((((((.....)))))))))....)))..))))-..))))))))))-----)((((.((....)).)-))).... ( -32.90) >DroSim_CAF1 16657 101 - 1 G-UGGGGGCGUGUCUCAAAUCGGCUGCCUAAAUAAGAAAUCAUUUUAUUUGAAAAGUUUCCCUUUGGCACGCCCCCUCCAGCUGCCGCCC-CACUUUU-CGAGG-- .-.(((((((((((.......((((...(((((((((.....)))))))))...)))).......)))))))))))....((....))..-.......-.....-- ( -31.34) >DroEre_CAF1 11918 102 - 1 A-UGGGGGCGUGGCUCAAAUCGGCUGCCUAAAUAAGAAAUCAUUUUAUUUGAAAAGUUUCCCUUCGGCACGCCCCCUCCAGCUGCCGCCCCCACUCUU-CGCUG-- .-((((((((..(((......((((((((((((((((.....)))))))))..(((.....))).)))).)))......)))..).))))))).....-.....-- ( -36.20) >DroYak_CAF1 12322 102 - 1 A-UGGGGGCGUGUCUCAAAUCGGCUGCCUAAAUAAGAAAUCAUUUUAUUUGAAAAGUUUCCCAUCGGCACGCCCCCUCCAGCUGCCGCCCCCACUUUU-CGCUG-- .-.(((((((((((.......((((...(((((((((.....)))))))))...)))).......)))))))))))..((((................-.))))-- ( -32.67) >DroPer_CAF1 13541 99 - 1 GGGAAGGGCGUGUCUCAAAUCGGAUGCCUAAAUAAGAAAUCAUUUUAUUUGAAAAGUUUCGUUU-UCCACGCCCCUU-----UGCCCUCGCCCCUGUG-GGCUGCG (((..(((((((....((((..(((...(((((((((.....)))))))))....)))..))))-..)))))))...-----..)))..((((....)-))).... ( -29.50) >consensus G_UGGGGGCGUGUCUCAAAUCGGCUGCCUAAAUAAGAAAUCAUUUUAUUUGAAAAGUUUCCCUU_GGCACGCCCCCUCCAGCUGCCGCCCCCACUUUU_CGCUG__ ...(((((((((.........((((...(((((((((.....)))))))))...)))).........))))))))).............................. (-22.06 = -22.04 + -0.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:07:23 2006