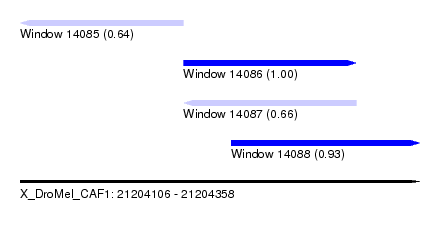
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 21,204,106 – 21,204,358 |
| Length | 252 |
| Max. P | 0.999443 |

| Location | 21,204,106 – 21,204,209 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.90 |
| Mean single sequence MFE | -26.23 |
| Consensus MFE | -18.08 |
| Energy contribution | -18.87 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.640976 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21204106 103 - 22224390 CAUAACUGCACCAGAGAACCCACCGACACUGUCCCAGAUCUUGGUGUCUUAUUCGAUCAUAGGAUCUGGAUCAAUGUAGAGCGAUCCAUAAGUAAUCCACCAA----------------- .....(((((..............(((...)))((((((((((..(((......)))..)))))))))).....)))))........................----------------- ( -21.10) >DroSec_CAF1 7790 116 - 1 ----UCUGCACUAGAAUCCCCCCGGAUAAAGUCCCAGAUCUUGGUGUCCUAUUCGACCAUAGGAUCUGGAUCAAUGUAGAGCGAUCCAUAAGUAAACCUUCAAUCUUAUACGUUGCCUUA ----((((((.....((((....))))...(..((((((((((..(((......)))..))))))))))..)..))))))(((((..(((((............)))))..))))).... ( -31.70) >DroSim_CAF1 4459 115 - 1 ----UCUGCACUAGAAUCCCUCCGGAUAAUGUCCCAGAUCUUUGUGUCUUAUUCCACCAUAGGAUCUGGAUCAAGGUGGAGCGAUCCAUAAGUAAACCUUCAAUC-UAUACGUUGCCUUA ----...((((((((((((....))))..((..(((((((((.(((........)))...)))))))))..))..(((((....)))))..............))-))...).))).... ( -25.90) >consensus ____UCUGCACUAGAAUCCCCCCGGAUAAUGUCCCAGAUCUUGGUGUCUUAUUCGACCAUAGGAUCUGGAUCAAUGUAGAGCGAUCCAUAAGUAAACCUUCAAUC_UAUACGUUGCCUUA .....((((......((((....))))...(..((((((((((..(((......)))..))))))))))..)...))))......................................... (-18.08 = -18.87 + 0.78)

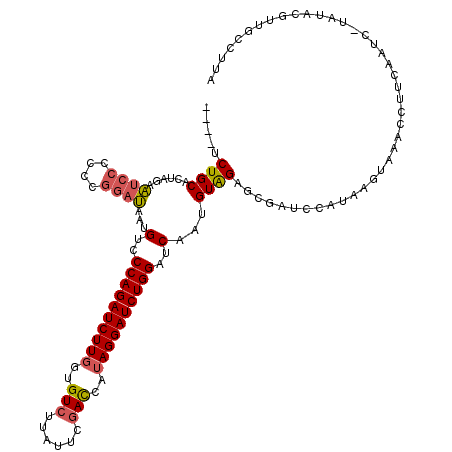

| Location | 21,204,209 – 21,204,318 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 79.08 |
| Mean single sequence MFE | -31.83 |
| Consensus MFE | -26.39 |
| Energy contribution | -26.83 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.52 |
| Structure conservation index | 0.83 |
| SVM decision value | 3.61 |
| SVM RNA-class probability | 0.999443 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21204209 109 + 22224390 UGCUAAUACGUCGCGAGCCUGAUCUAAUCAGUAUUAAGCAGUAGCUUCUUGGCUU-GGCAUCAGGAACGAUGGUUAUUGAAGUCGAGCUGGAGCUGCUUGUAGAAAUUAA ((((((((((.(....).)((((...))))))))).))))((((((((..(((((-(((.((((.(((....))).)))).))))))))))))))))............. ( -35.90) >DroSec_CAF1 7906 86 + 1 ------------------------UAAUCAGUAUUAAGCAGUAGCUUCUUGGCUUUGCCAUCAGUAUCGAUGGUUAUUGAAGUCAAGCUGGGGCUGCUUGUAGAAAUUAU ------------------------..........((((((((..(..(((((((((((((((......))))))....)))))))))..)..)))))))).......... ( -29.70) >DroSim_CAF1 4574 86 + 1 ------------------------UAAUCAGUAUUAAGCAGUAGCUUCUUGGCUUUGCCAUCAGUAUCGAUGGUUAUUGAAGUCAAGCUGGGGCUGCUUGCAGAAAUUAU ------------------------.............(((((((((((((((((((((((((......))))))....)))))))))...))))))).)))......... ( -29.90) >consensus ________________________UAAUCAGUAUUAAGCAGUAGCUUCUUGGCUUUGCCAUCAGUAUCGAUGGUUAUUGAAGUCAAGCUGGGGCUGCUUGUAGAAAUUAU ..................................((((((((..(..(((((((((((((((......))))))....)))))))))..)..)))))))).......... (-26.39 = -26.83 + 0.45)



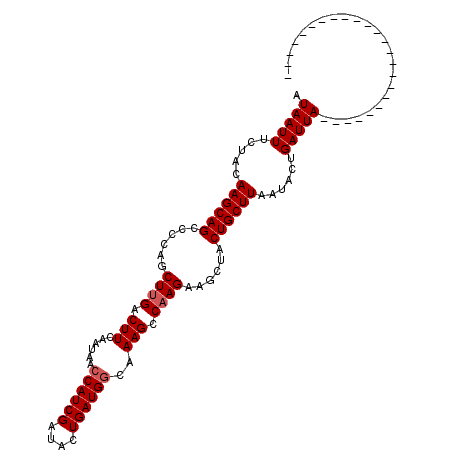
| Location | 21,204,209 – 21,204,318 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 79.08 |
| Mean single sequence MFE | -17.70 |
| Consensus MFE | -14.83 |
| Energy contribution | -15.50 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.664542 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21204209 109 - 22224390 UUAAUUUCUACAAGCAGCUCCAGCUCGACUUCAAUAACCAUCGUUCCUGAUGCC-AAGCCAAGAAGCUACUGCUUAAUACUGAUUAGAUCAGGCUCGCGACGUAUUAGCA .............(((((....))).......((((....((((.((((((...-.((.....((((....))))....))......))))))...))))..)))).)). ( -18.10) >DroSec_CAF1 7906 86 - 1 AUAAUUUCUACAAGCAGCCCCAGCUUGACUUCAAUAACCAUCGAUACUGAUGGCAAAGCCAAGAAGCUACUGCUUAAUACUGAUUA------------------------ .(((((.....((((((......((((.(((......((((((....))))))..))).))))......))))))......)))))------------------------ ( -17.50) >DroSim_CAF1 4574 86 - 1 AUAAUUUCUGCAAGCAGCCCCAGCUUGACUUCAAUAACCAUCGAUACUGAUGGCAAAGCCAAGAAGCUACUGCUUAAUACUGAUUA------------------------ .(((((.....((((((......((((.(((......((((((....))))))..))).))))......))))))......)))))------------------------ ( -17.50) >consensus AUAAUUUCUACAAGCAGCCCCAGCUUGACUUCAAUAACCAUCGAUACUGAUGGCAAAGCCAAGAAGCUACUGCUUAAUACUGAUUA________________________ .(((((.....((((((......((((.(((......((((((....))))))..))).))))......))))))......)))))........................ (-14.83 = -15.50 + 0.67)


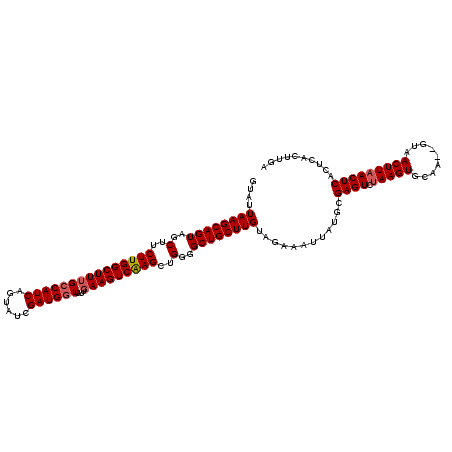
| Location | 21,204,239 – 21,204,358 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.18 |
| Mean single sequence MFE | -37.83 |
| Consensus MFE | -32.62 |
| Energy contribution | -33.40 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925386 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21204239 119 + 22224390 GUAUUAAGCAGUAGCUUCUUGGCUU-GGCAUCAGGAACGAUGGUUAUUGAAGUCGAGCUGGAGCUGCUUGUAGAAAUUAAGCGAGUCUAAGUGCAUACACAACUUAACUCACUCACUUGA .......(((((((((((..(((((-(((.((((.(((....))).)))).)))))))))))))))).))).....(((((.((((.(((((.........)))))....)))).))))) ( -38.70) >DroSec_CAF1 7912 118 + 1 GUAUUAAGCAGUAGCUUCUUGGCUUUGCCAUCAGUAUCGAUGGUUAUUGAAGUCAAGCUGGGGCUGCUUGUAGAAAUUAUGCGAGUCUAAGUGCAA--GUAACUUAACUCACUCACUUGA ((((((((((((..(..(((((((((((((((......))))))....)))))))))..)..))))))))........))))((((.(((((....--...))))))))).......... ( -37.80) >DroSim_CAF1 4580 118 + 1 GUAUUAAGCAGUAGCUUCUUGGCUUUGCCAUCAGUAUCGAUGGUUAUUGAAGUCAAGCUGGGGCUGCUUGCAGAAAUUAUGCGAGUCAAAGUGCAA--GUAACUUAACUCACUCACUUGA .....(((((((..(..(((((((((((((((......))))))....)))))))))..)..)))))))(((.......)))((((..((((....--...)))).)))).......... ( -37.00) >consensus GUAUUAAGCAGUAGCUUCUUGGCUUUGCCAUCAGUAUCGAUGGUUAUUGAAGUCAAGCUGGGGCUGCUUGUAGAAAUUAUGCGAGUCUAAGUGCAA__GUAACUUAACUCACUCACUUGA ....((((((((..(..(((((((((((((((......))))))....)))))))))..)..))))))))............((((.(((((.........))))))))).......... (-32.62 = -33.40 + 0.78)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:07:21 2006