
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 21,181,873 – 21,181,984 |
| Length | 111 |
| Max. P | 0.878132 |

| Location | 21,181,873 – 21,181,984 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.31 |
| Mean single sequence MFE | -40.50 |
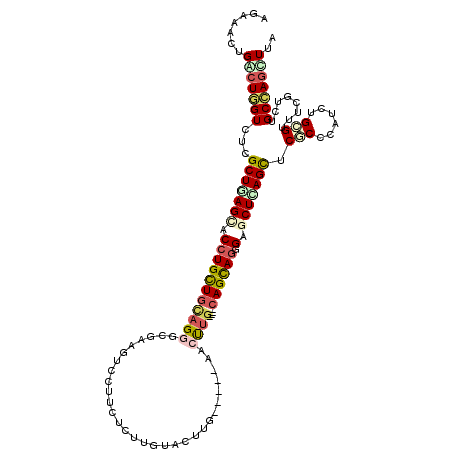
| Consensus MFE | -20.28 |
| Energy contribution | -21.31 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.878132 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
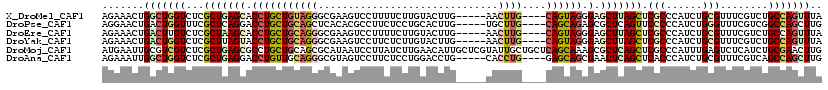
>X_DroMel_CAF1 21181873 111 - 22224390 AGAAACUGGCUGGUCUCGCUGAGCACCUGCUGUAGGGCGAAGUCCUUUUCUUGUACUUG-----AACUUG----CAGUAGGGAGCUUAGCUCGCCCAUCUGCGUUUCGUCUGCCAGUUUA ..((((((((.((.(..(((((((.(((((((((((...((((.(.......).)))).-----..))))----)))))))..))))))).(((......)))....).)))))))))). ( -46.60) >DroPse_CAF1 400 111 - 1 AGGAACUGACUGGUUUCGCUCAGGACCUGCUGCAGCUCACACGCCUUCUCCUGCACUUG-----UGCUUG----CAGCAGAGCGCUCAGUUCCCCCAUCUGGGUUUCGUCGGCCAGCUUG .((((((((((((......))))(..((((((((((.((...((........))...))-----.)).))----))))))..)..))))))))(((....)))................. ( -37.20) >DroEre_CAF1 8188 111 - 1 AGAAACUGACUUGUCUCGCUAAGCACCUGCUGCAGGGCGAAGUCCUUUUCUUGUACUUG-----AACUUG----CAGUAGGGAGCUUAGCUCGCCCAUCUGCGUUUCGUCUGCCAGUUUA ..((((((.........(((((((.(((((((((((...((((.(.......).)))).-----..))))----)))))))..))))))).(((......)))..........)))))). ( -41.30) >DroYak_CAF1 1394 111 - 1 AGAAACUGACUGGUCUCGCUUAGUACCUGCUGCAGGGCGAAGUCCUUCUCUUGUACUUG-----AACUUG----CAGUAGGGAGCUUAGCUCGCCCAUCUGCGUUUCGUCUGCCAGUUUA .......(((((((...(((.(((.(((((((((((...((((.(.......).)))).-----..))))----)))))))..))).))).(((......)))........))))))).. ( -38.30) >DroMoj_CAF1 8689 120 - 1 AUGAAUUGCGUCGUCUCGCUGAGCGCCUGCUGCAGCGCAUAAUCCUUAUCUUGAACAUUGCUCGUAUUGCUGCUCAGCAAAGCGCUCAGCUCGCCCAUUUGAGUCUCAUCUGCGAACUUG .................(((((((((.(((((.((((((............................))).))))))))..)))))))))((((.....((.....))...))))..... ( -34.99) >DroAna_CAF1 7314 111 - 1 AGAAAUUGGCUGGUCUCGCUGAGGACCUGUUGCAGGGCGUAGUCCUUCUCCUGGACCUG-----CACCUG----GAGCAGCGAACUCAGCUCACCCAUCUGCGUUUCGUCAGCCAGCUUG .......((((((....((((((..((((((.((((..(((((((.......))).)))-----).))))----.))))).)..))))))........(((((...)).))))))))).. ( -44.60) >consensus AGAAACUGACUGGUCUCGCUGAGCACCUGCUGCAGGGCGAAGUCCUUCUCUUGUACUUG_____AACUUG____CAGCAGGGAGCUCAGCUCGCCCAUCUGCGUUUCGUCUGCCAGCUUA .......(((((((...(((((((.(((((((((((..............................))))....)))))).).))))))).(((......)))........))))))).. (-20.28 = -21.31 + 1.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:07:11 2006